 |
PDBsum entry 1ndf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.3.1.137
- carnitine O-octanoyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
octanoyl-CoA + (R)-carnitine = O-octanoyl-(R)-carnitine + CoA
|
 |
 |
 |
 |
 |
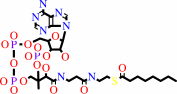 octanoyl-CoA
octanoyl-CoA
|
+
|
(R)-carnitine
|
=
|
O-octanoyl-(R)-carnitine
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.3.1.7
- carnitine O-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-carnitine + acetyl-CoA = O-acetyl-(R)-carnitine + CoA
|
 |
 |
 |
 |
 |
(R)-carnitine
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
O-acetyl-(R)-carnitine
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cell
112:113-122
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of carnitine acetyltransferase and implications for the catalytic mechanism and fatty acid transport.
|
|
G.Jogl,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Carnitine acyltransferases have crucial roles in the transport of fatty acids
for beta-oxidation. Dysregulation of these enzymes can lead to serious diseases
in humans, and they are targets for therapeutic development against diabetes. We
report the crystal structures of murine carnitine acetyltransferase (CRAT),
alone and in complex with its substrate carnitine or CoA. The structure contains
two domains. Surprisingly, these two domains share the same backbone fold, which
is also similar to that of chloramphenicol acetyltransferase and dihydrolipoyl
transacetylase. The active site is located at the interface between the two
domains. Carnitine and CoA are bound in deep channels in the enzyme, on opposite
sides of the catalytic His343 residue. The structural information provides a
molecular basis for understanding the catalysis by carnitine acyltransferases
and for designing their inhibitors. Specifically, our structural information
suggests that the substrate carnitine may assist the catalysis by stabilizing
the oxyanion in the reaction intermediate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. The CoA Binding Site of CRAT(A) Final
2F[o]–F[c] electron density map for CoA at 2.3 Å
resolution. The contour level is at 1σ. Produced with Setor
(Evans, 1993).(B) Stereo diagram showing the CoA binding site of
CRAT. The CoA molecule is shown in brown. Produced with
Ribbons (Carson, 1987).(C) Overlap of the binding modes of CoA
to CRAT (in brown) and CAT (in cyan).(D) Molecular surface of
CRAT in the region of the CoA binding site. (C and D) produced
with Grasp (Nicholls et al., 1991).
|
 |
Figure 6.
Figure 6. The Catalytic Mechanism of Carnitine
AcyltransferasesThe catalytic His343 residue can extract the
proton from either carnitine or CoA. The oxyanion in the
tetrahedral intermediate is stabilized by interactions with
carnitine and the side chain hydroxyl of Ser554.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Cell
(2003,
112,
113-122)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Morar,
and
G.D.Wright
(2010).
The genomic enzymology of antibiotic resistance.
|
| |
Annu Rev Genet,
44,
25-51.
|
 |
|
|
|
|
 |
N.T.Price,
V.N.Jackson,
J.Müller,
K.Moffat,
K.L.Matthews,
T.Orton,
and
V.A.Zammit
(2010).
Alternative exon usage in the single CPT1 gene of Drosophila generates functional diversity in the kinetic properties of the enzyme: differential expression of alternatively spliced variants in Drosophila tissues.
|
| |
J Biol Chem,
285,
7857-7865.
|
 |
|
|
|
|
 |
A.C.Rufer,
R.Thoma,
and
M.Hennig
(2009).
Structural insight into function and regulation of carnitine palmitoyltransferase.
|
| |
Cell Mol Life Sci,
66,
2489-2501.
|
 |
|
|
|
|
 |
N.Shah,
S.Khurana,
K.Cheng,
and
J.P.Raufman
(2009).
Muscarinic receptors and ligands in cancer.
|
| |
Am J Physiol Cell Physiol,
296,
C221-C232.
|
 |
|
|
|
|
 |
T.Y.Hou,
S.M.Ward,
J.M.Murad,
N.P.Watson,
M.A.Israel,
and
G.E.Duffield
(2009).
ID2 (inhibitor of DNA binding 2) is a rhythmically expressed transcriptional repressor required for circadian clock output in mouse liver.
|
| |
J Biol Chem,
284,
31735-31745.
|
 |
|
|
|
|
 |
Y.Kim,
H.Li,
T.A.Binkowski,
D.Holzle,
and
A.Joachimiak
(2009).
Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis.
|
| |
J Struct Funct Genomics,
10,
157-163.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Wolfgang,
S.H.Cha,
D.S.Millington,
G.Cline,
G.I.Shulman,
A.Suwa,
M.Asaumi,
T.Kurama,
T.Shimokawa,
and
M.D.Lane
(2008).
Brain-specific carnitine palmitoyl-transferase-1c: role in CNS fatty acid metabolism, food intake, and body weight.
|
| |
J Neurochem,
105,
1550-1559.
|
 |
|
|
|
|
 |
A.Faye,
C.Esnous,
N.T.Price,
M.A.Onfray,
J.Girard,
and
C.Prip-Buus
(2007).
Rat liver carnitine palmitoyltransferase 1 forms an oligomeric complex within the outer mitochondrial membrane.
|
| |
J Biol Chem,
282,
26908-26916.
|
 |
|
|
|
|
 |
C.Bolduc,
M.Yoshioka,
and
J.St-Amand
(2007).
Transcriptomic characterization of the long-term dihydrotestosterone effects in adipose tissue.
|
| |
Obesity (Silver Spring),
15,
1107-1132.
|
 |
|
|
|
|
 |
H.Unno,
F.Ichimaida,
H.Suzuki,
S.Takahashi,
Y.Tanaka,
A.Saito,
T.Nishino,
M.Kusunoki,
and
T.Nakayama
(2007).
Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis.
|
| |
J Biol Chem,
282,
15812-15822.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Chiechio,
A.Copani,
R.W.Gereau,
and
F.Nicoletti
(2007).
Acetyl-L-carnitine in neuropathic pain: experimental data.
|
| |
CNS Drugs,
21,
31.
|
 |
|
|
|
|
 |
K.Borthwick,
V.N.Jackson,
N.T.Price,
and
V.A.Zammit
(2006).
The mitochondrial intermembrane loop region of rat carnitine palmitoyltransferase 1A is a major determinant of its malonyl-CoA sensitivity.
|
| |
J Biol Chem,
281,
32946-32952.
|
 |
|
|
|
|
 |
M.J.Wolfgang,
T.Kurama,
Y.Dai,
A.Suwa,
M.Asaumi,
S.Matsumoto,
S.H.Cha,
T.Shimokawa,
and
M.D.Lane
(2006).
The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis.
|
| |
Proc Natl Acad Sci U S A,
103,
7282-7287.
|
 |
|
|
|
|
 |
S.Chiechio,
A.Copani,
F.Nicoletti,
and
R.Gereau Iv
(2006).
L-acetylcarnitine: a proposed therapeutic agent for painful peripheral neuropathies.
|
| |
Curr Neuropharmacol,
4,
233-237.
|
 |
|
|
|
|
 |
Y.S.Hsiao,
G.Jogl,
and
L.Tong
(2006).
Crystal structures of murine carnitine acetyltransferase in ternary complexes with its substrates.
|
| |
J Biol Chem,
281,
28480-28487.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.S.Hsiao,
G.Jogl,
V.Esser,
and
L.Tong
(2006).
Crystal structure of rat carnitine palmitoyltransferase II (CPT-II).
|
| |
Biochem Biophys Res Commun,
346,
974-980.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.R.Kim,
T.Dobransky,
R.J.Rylett,
and
B.H.Shilton
(2005).
Surface-entropy reduction used in the crystallization of human choline acetyltransferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1306-1310.
|
 |
|
|
|
|
 |
G.Jogl,
Y.S.Hsiao,
and
L.Tong
(2005).
Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
|
| |
J Biol Chem,
280,
738-744.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Liu,
G.Zheng,
M.Treber,
J.Dai,
and
G.Woldegiorgis
(2005).
Cysteine-scanning mutagenesis of muscle carnitine palmitoyltransferase I reveals a single cysteine residue (Cys-305) is important for catalysis.
|
| |
J Biol Chem,
280,
4524-4531.
|
 |
|
|
|
|
 |
K.C.Onwueme,
C.J.Vos,
J.Zurita,
J.A.Ferreras,
and
L.E.Quadri
(2005).
The dimycocerosate ester polyketide virulence factors of mycobacteria.
|
| |
Prog Lipid Res,
44,
259-302.
|
 |
|
|
|
|
 |
R.D.Gandour
(2005).
Rationalizing the solution properties of zwitterions by means of computational chemistry.
|
| |
Chem Biodivers,
2,
1580-1594.
|
 |
|
|
|
|
 |
X.Ma,
J.Koepke,
S.Panjikar,
G.Fritzsch,
and
J.Stöckigt
(2005).
Crystal structure of vinorine synthase, the first representative of the BAHD superfamily.
|
| |
J Biol Chem,
280,
13576-13583.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.G.Cordente,
E.López-Viñas,
M.I.Vázquez,
J.H.Swiegers,
I.S.Pretorius,
P.Gómez-Puertas,
F.G.Hegardt,
G.Asins,
and
D.Serra
(2004).
Redesign of carnitine acetyltransferase specificity by protein engineering.
|
| |
J Biol Chem,
279,
33899-33908.
|
 |
|
|
|
|
 |
G.Jogl,
Y.S.Hsiao,
and
L.Tong
(2004).
Structure and function of carnitine acyltransferases.
|
| |
Ann N Y Acad Sci,
1033,
17-29.
|
 |
|
|
|
|
 |
J.Buglino,
K.C.Onwueme,
J.A.Ferreras,
L.E.Quadri,
and
C.D.Lima
(2004).
Crystal structure of PapA5, a phthiocerol dimycocerosyl transferase from Mycobacterium tuberculosis.
|
| |
J Biol Chem,
279,
30634-30642.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Lian,
Y.Gu,
B.Pedersen,
T.Kukar,
L.Govindasamy,
M.Agbandje-McKenna,
S.Jin,
R.McKenna,
and
D.Wu
(2004).
Crystallization and preliminary X-ray crystallographic studies on recombinant rat choline acetyltransferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
374-375.
|
 |
|
|
|
|
 |
Y.Cai,
C.N.Cronin,
A.G.Engel,
K.Ohno,
L.B.Hersh,
and
D.W.Rodgers
(2004).
Choline acetyltransferase structure reveals distribution of mutations that cause motor disorders.
|
| |
EMBO J,
23,
2047-2058.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.S.Hsiao,
G.Jogl,
and
L.Tong
(2004).
Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase.
|
| |
J Biol Chem,
279,
31584-31589.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Shi,
and
P.Burn
(2004).
Lipid metabolic enzymes: emerging drug targets for the treatment of obesity.
|
| |
Nat Rev Drug Discov,
3,
695-710.
|
 |
|
|
|
|
 |
L.Napal,
J.Dai,
M.Treber,
D.Haro,
P.F.Marrero,
and
G.Woldegiorgis
(2003).
A single amino acid change (substitution of the conserved Glu-590 with alanine) in the C-terminal domain of rat liver carnitine palmitoyltransferase I increases its malonyl-CoA sensitivity close to that observed with the muscle isoform of the enzyme.
|
| |
J Biol Chem,
278,
34084-34089.
|
 |
|
|
|
|
 |
M.Valle,
R.Gillet,
S.Kaur,
A.Henne,
V.Ramakrishnan,
and
J.Frank
(2003).
Visualizing tmRNA entry into a stalled ribosome.
|
| |
Science,
300,
127-130.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.R.Ramsay,
and
J.H.Naismith
(2003).
A snapshot of carnitine acetyltransferase.
|
| |
Trends Biochem Sci,
28,
343-346.
|
 |
|
|
|
|
 |
S.Gobin,
L.Thuillier,
G.Jogl,
A.Faye,
L.Tong,
M.Chi,
J.P.Bonnefont,
J.Girard,
and
C.Prip-Buus
(2003).
Functional and structural basis of carnitine palmitoyltransferase 1A deficiency.
|
| |
J Biol Chem,
278,
50428-50434.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

