 |
PDBsum entry 1kh2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.5
- argininosuccinate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-citrulline + L-aspartate + ATP = 2-(N(omega)-L-arginino)succinate + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
L-citrulline
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 L-aspartate
L-aspartate
|
+
|
 ATP
ATP
|
=
|
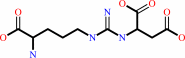 2-(N(omega)-L-arginino)succinate
2-(N(omega)-L-arginino)succinate
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:15890-15896
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of argininosuccinate synthetase from Thermus thermophilus HB8. Structural basis for the catalytic action.
|
|
M.Goto,
Y.Nakajima,
K.Hirotsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Argininosuccinate synthetase catalyzes the ATP-dependent condensation of a
citrulline with an aspartate to give argininosuccinate. The three-dimensional
structures of the enzyme from Thermus thermophilus HB8 in its free form,
complexed with intact ATP, and complexed with an ATP analogue (adenylyl
imidodiphosphate) and substrate analogues (arginine and succinate) have been
determined at 2.3-, 2.3-, and 1.95-A resolution, respectively. The structure is
essentially the same as that of the Escherichia coli argininosuccinate
synthetase. The small domain has the same fold as that of a new family of
"N-type" ATP pyrophosphatases with the P-loop specific for the
pyrophosphate of ATP. However, the enzyme shows the P-loop specific for the
gamma-phosphate of ATP. The structure of the complex form is quite similar to
that of the native one, indicating that no conformational change occurs upon the
binding of ATP and the substrate analogues. ATP and the substrate analogues are
bound to the active site with their reaction sites close to one another and
located in a geometrical orientation favorable to the catalytic action. The
reaction mechanism so far proposed seems to be consistent with the locations of
ATP and the substrate analogues. The reaction may proceed without the large
conformational change of the enzyme proposed for the catalytic process.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. Schematic diagram showing hydrogen bond and salt
bridge interactions of the active site residues in
tAsS·AMP-PNP·arginine·succinate. Putative
interactions are shown by dotted lines if the acceptor and donor
are less than 3.5 Å apart. W indicates a water molecule.
AMP-PNP, arginine, and succinate bound to the active site are
drawn by thick bonds.
|
 |
Figure 5.
Fig. 5. Model of ATP, citrulline, and aspartate binding
to tAsS. Important short contacts are shown by dotted lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
15890-15896)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.F.Gherardini,
G.Ausiello,
and
M.Helmer-Citterich
(2010).
Superpose3D: a local structural comparison program that allows for user-defined structure representations.
|
| |
PLoS One,
5,
e11988.
|
 |
|
|
|
|
 |
T.Karlberg,
R.Collins,
S.van den Berg,
A.Flores,
M.Hammarström,
M.Högbom,
L.Holmberg Schiavone,
and
J.Uppenberg
(2008).
Structure of human argininosuccinate synthetase.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
279-286.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kuratani,
Y.Yoshikawa,
Y.Bessho,
K.Higashijima,
T.Ishii,
R.Shibata,
S.Takahashi,
K.Yutani,
and
S.Yokoyama
(2007).
Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine.
|
| |
Structure,
15,
1642-1653.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Curis,
I.Nicolis,
C.Moinard,
S.Osowska,
N.Zerrouk,
S.Bénazeth,
and
L.Cynober
(2005).
Almost all about citrulline in mammals.
|
| |
Amino Acids,
29,
177-205.
|
 |
|
|
|
|
 |
A.Husson,
C.Brasse-Lagnel,
A.Fairand,
S.Renouf,
and
A.Lavoinne
(2003).
Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle.
|
| |
Eur J Biochem,
270,
1887-1899.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

