 |
PDBsum entry 1ilo

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics
|
PDB id
|
|

|

|
1ilo
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.4.8
- phosphoadenylyl-sulfate reductase (thioredoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[thioredoxin]-disulfide + sulfite + adenosine 3',5'-bisphosphate + 2 H+ = [thioredoxin]-dithiol + 3'-phosphoadenylyl sulfate
|
 |
 |
 |
 |
 |
[thioredoxin]-disulfide
|
+
|
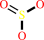 sulfite
sulfite
|
+
|
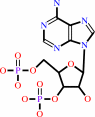 adenosine 3',5'-bisphosphate
adenosine 3',5'-bisphosphate
|
+
|
2
×
H(+)
|
=
|
[thioredoxin]-dithiol
|
+
|
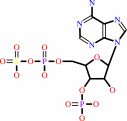 3'-phosphoadenylyl sulfate
3'-phosphoadenylyl sulfate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:4760-4770
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification of a novel archaebacterial thioredoxin: determination of function through structure.
|
|
S.Bhattacharyya,
B.Habibi-Nazhad,
G.Amegbey,
C.M.Slupsky,
A.Yee,
C.Arrowsmith,
D.S.Wishart.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
As part of a high-throughput, structural proteomic project we have used NMR
spectroscopy to determine the solution structure and ascertain the function of a
previously unknown, conserved protein (MtH895) from the thermophilic archeon
Methanobacterium thermoautotrophicum. Our findings indicate that MtH895 contains
a central four-stranded beta-sheet core surrounded by two helices on one side
and a third on the other. It has an overall fold superficially similar to that
of a glutaredoxin. However, detailed analysis of its three-dimensional structure
along with molecular docking simulations of its interaction with T7 DNA
polymerase (a thioredoxin-specific substrate) and comparisons with other known
members of the thioredoxin/glutaredoxin family of proteins strongly suggest that
MtH895 is more akin to a thioredoxin. Furthermore, measurement of the pK(a)
values of its active site thiols along with direct measurements of the
thioredoxin/glutaredoxin activity has confirmed that MtH895 is, indeed, a
thioredoxin and exhibits no glutaredoxin activity. We have also identified a
group of previously unknown proteins from several other archaebacteria that have
significant (34-44%) sequence identity with MtH895. These proteins have unusual
active site -CXXC- motifs not found in any known thioredoxin or glutaredoxin. On
the basis of the results presented here, we predict that these small proteins
are all members of a new class of truncated thioredoxins.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Carella,
J.Becher,
O.Ohlenschläger,
R.Ramachandran,
K.H.Gührs,
G.Wellenreuther,
W.Meyer-Klaucke,
S.H.Heinemann,
and
M.Görlach
(2011).
Structure-function relationship in an archaebacterial methionine sulphoxide reductase B.
|
| |
Mol Microbiol,
79,
342-358.
|
 |
|
|
|
|
 |
E.Pedone,
D.Limauro,
and
S.Bartolucci
(2008).
The machinery for oxidative protein folding in thermophiles.
|
| |
Antioxid Redox Signal,
10,
157-170.
|
 |
|
|
|
|
 |
C.R.Guzzo,
R.A.Nagem,
L.M.Galvão-Botton,
B.G.Guimarães,
F.J.Medrano,
J.A.Barbosa,
and
C.S.Farah
(2005).
Expression, purification, crystallization and preliminary X-ray analysis of YaeQ (XAC2396) from Xanthomonas axonopodis pv. citri.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
493-495.
|
 |
|
|
|
|
 |
J.Eichler,
and
M.W.Adams
(2005).
Posttranslational protein modification in Archaea.
|
| |
Microbiol Mol Biol Rev,
69,
393-425.
|
 |
|
|
|
|
 |
J.Wang,
T.Wang,
E.R.Zuiderweg,
and
G.M.Crippen
(2005).
CASA: an efficient automated assignment of protein mainchain NMR data using an ordered tree search algorithm.
|
| |
J Biomol NMR,
33,
261-279.
|
 |
|
|
|
|
 |
A.F.Yakunin,
A.A.Yee,
A.Savchenko,
A.M.Edwards,
and
C.H.Arrowsmith
(2004).
Structural proteomics: a tool for genome annotation.
|
| |
Curr Opin Chem Biol,
8,
42-48.
|
 |
|
|
|
|
 |
E.Pedone,
B.Ren,
R.Ladenstein,
M.Rossi,
and
S.Bartolucci
(2004).
Functional properties of the protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus: a member of a novel protein family related to protein disulfide-isomerase.
|
| |
Eur J Biochem,
271,
3437-3448.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

