 |
PDBsum entry 1gpu

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.1
- transketolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = aldehydo-D- ribose 5-phosphate + D-xylulose 5-phosphate
|
 |
 |
 |
 |
 |
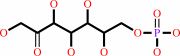 D-sedoheptulose 7-phosphate
D-sedoheptulose 7-phosphate
|
+
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
=
|
aldehydo-D- ribose 5-phosphate
|
+
|
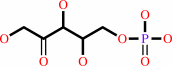 D-xylulose 5-phosphate
D-xylulose 5-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
THD)
matches with 86.67% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
99:591-595
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Snapshot of a key intermediate in enzymatic thiamin catalysis: crystal structure of the alpha-carbanion of (alpha,beta-dihydroxyethyl)-thiamin diphosphate in the active site of transketolase from Saccharomyces cerevisiae.
|
|
E.Fiedler,
S.Thorell,
T.Sandalova,
R.Golbik,
S.König,
G.Schneider.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Kinetic and spectroscopic data indicated that addition of the donor substrate
hydroxypyruvate to the thiamin diphosphate (ThDP)-dependent enzyme transketolase
(TK) led to the accumulation of the alpha-carbanion/enamine of
(alpha,beta-dihydroxyethyl) ThDP, the key reaction intermediate in enzymatic
thiamin catalysis. The three-dimensional structure of this intermediate trapped
in the active site of yeast TK was determined to 1.9-A resolution by using
cryocrystallography. The electron density suggests a planar
alpha-carbanion/enamine intermediate having the E-configuration. The reaction
intermediate is firmly held in place through direct hydrogen bonds to His-103
and His-481 and an indirect hydrogen bond via a water molecule to His-69. The
4-NH(2) group of the amino-pyrimidine ring of ThDP is within 3 A distance to the
alpha-hydroxy oxygen atom of the dihydroxyethyl moiety but at an angle
unfavorable for a strong hydrogen bond. No structural changes occur in TK on
formation of the reaction intermediate, suggesting that the active site is
poised for catalysis and conformational changes during the enzyme reaction are
not very likely. The intermediate is present with high occupancy in both active
sites, arguing against previous proposals of half-of-the-sites reactivity in
yeast TK.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Formation of the  -carbanion/enamine
intermediate by using HPA as donor substrate. -carbanion/enamine
intermediate by using HPA as donor substrate.
|
 |
Figure 5.
Fig. 5. Superposition of the ThDP molecule bound in
holoTK with the covalent reaction intermediate DHEThDP in the
TK-intermediate complex. The superposition is based on 678 C
 atoms of
the TK subunit. atoms of
the TK subunit.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Cázares,
J.L.Galman,
L.G.Crago,
M.E.Smith,
J.Strafford,
L.Ríos-Solís,
G.J.Lye,
P.A.Dalby,
and
H.C.Hailes
(2010).
Non-alpha-hydroxylated aldehydes with evolved transketolase enzymes.
|
| |
Org Biomol Chem,
8,
1301-1309.
|
 |
|
|
|
|
 |
M.Koutmos,
O.Kabil,
J.L.Smith,
and
R.Banerjee
(2010).
Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine {beta}-synthase.
|
| |
Proc Natl Acad Sci U S A,
107,
20958-20963.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Fushinobu
(2010).
Unique sugar metabolic pathways of bifidobacteria.
|
| |
Biosci Biotechnol Biochem,
74,
2374-2384.
|
 |
|
|
|
|
 |
L.E.Meshalkina,
G.A.Kochetov,
G.Hübner,
K.Tittmann,
and
R.Golbik
(2009).
New function of the amino group of thiamine diphosphate in thiamine catalysis.
|
| |
Biochemistry (Mosc),
74,
293-300.
|
 |
|
|
|
|
 |
A.Yep,
G.L.Kenyon,
and
M.J.McLeish
(2008).
Saturation mutagenesis of putative catalytic residues of benzoylformate decarboxylase provides a challenge to the accepted mechanism.
|
| |
Proc Natl Acad Sci U S A,
105,
5733-5738.
|
 |
|
|
|
|
 |
S.J.Costelloe,
J.M.Ward,
and
P.A.Dalby
(2008).
Evolutionary Analysis of the TPP-Dependent Enzyme Family.
|
| |
J Mol Evol,
66,
36-49.
|
 |
|
|
|
|
 |
H.Xie,
S.Vucetic,
L.M.Iakoucheva,
C.J.Oldfield,
A.K.Dunker,
Z.Obradovic,
and
V.N.Uversky
(2007).
Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins.
|
| |
J Proteome Res,
6,
1917-1932.
|
 |
|
|
|
|
 |
A.Baykal,
S.Chakraborty,
A.Dodoo,
and
F.Jordan
(2006).
Synthesis with good enantiomeric excess of both enantiomers of alpha-ketols and acetolactates by two thiamin diphosphate-dependent decarboxylases.
|
| |
Bioorg Chem,
34,
380-393.
|
 |
|
|
|
|
 |
G.Wille,
D.Meyer,
A.Steinmetz,
E.Hinze,
R.Golbik,
and
K.Tittmann
(2006).
The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
|
| |
Nat Chem Biol,
2,
324-328.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Bourgeois,
and
A.Royant
(2005).
Advances in kinetic protein crystallography.
|
| |
Curr Opin Struct Biol,
15,
538-547.
|
 |
|
|
|
|
 |
O.A.Esakova,
E.A.Khanova,
L.E.Meshalkina,
R.Golbik,
G.Hübner,
and
G.A.Kochetov
(2005).
Effect of transketolase substrates on holoenzyme reconstitution and stability.
|
| |
Biochemistry (Mosc),
70,
770-776.
|
 |
|
|
|
|
 |
R.Golbik,
L.E.Meshalkina,
T.Sandalova,
K.Tittmann,
E.Fiedler,
H.Neef,
S.König,
R.Kluger,
G.A.Kochetov,
G.Schneider,
and
G.Hübner
(2005).
Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae.
|
| |
FEBS J,
272,
1326-1342.
|
 |
|
|
|
|
 |
M.V.Kovina,
A.De Kok,
I.A.Sevostyanova,
L.S.Khailova,
N.V.Belkina,
and
G.A.Kochetov
(2004).
The molecular origin of the thiamine diphosphate-induced spectral bands of ThDP-dependent enzymes.
|
| |
Proteins,
56,
338-345.
|
 |
|
|
|
|
 |
O.A.Esakova,
L.E.Meshalkina,
R.Golbik,
G.Hübner,
and
G.A.Kochetov
(2004).
Donor substrate regulation of transketolase.
|
| |
Eur J Biochem,
271,
4189-4194.
|
 |
|
|
|
|
 |
A.Schütz,
T.Sandalova,
S.Ricagno,
G.Hübner,
S.König,
and
G.Schneider
(2003).
Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid.
|
| |
Eur J Biochem,
270,
2312-2321.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

