 |
PDBsum entry 1glv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Glutathione biosynthesis ligase
|
PDB id
|
|

|

|
1glv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.3
- glutathione synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
gamma-L-glutamyl-L-cysteine + glycine + ATP = glutathione + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
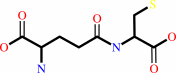 gamma-L-glutamyl-L-cysteine
gamma-L-glutamyl-L-cysteine
|
+
|
 glycine
glycine
|
+
|
 ATP
ATP
|
=
|
 glutathione
glutathione
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
229:1083-1100
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of the glutathione synthetase from Escherichia coli B at 2.0 A resolution.
|
|
H.Yamaguchi,
H.Kato,
Y.Hata,
T.Nishioka,
A.Kimura,
J.Oda,
Y.Katsube.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glutathione synthetase (gamma-L-glutamyl-L-cysteine: glycine ligase
(ADP-forming) EC 6.3.2.3: GSHase) catalyzes the synthesis of glutathione from
gamma-L-glutamyl-L-cysteine and Gly in the presence of ATP. The enzyme from
Escherichia coli is a tetramer with four identical subunits of 316 amino acid
residues. The crystal structure of the enzyme has been determined by isomorphous
replacement and refined to a 2.0 A resolution. Two regions, Gly164 to Gly167 and
Ile226 to Arg241, are invisible on the electron density map. The refined model
of the subunit includes 296 amino acid residues and 107 solvent molecules. The
crystallographic R-factor is 18.6% for 17.914 reflections with F > 3 sigma
between 6.0 A and 2.0 A. The structure consists of three domains: the
N-terminal, central, and C-terminal domains. In the tetrameric molecule, two
subunits that are in close contact form a tight dimer, two tight dimers forming
a tetramer with two solvent regions. The ATP molecule is located in the cleft
between the central and C-terminal domains. The ATP binding site is surrounded
by two sets of the structural motif that belong to those respective domains.
Each motif consists of an anti-parallel beta-sheet and a glycine-rich loop.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.K.Fyfe,
M.S.Alphey,
and
W.N.Hunter
(2010).
Structure of Trypanosoma brucei glutathione synthetase: domain and loop alterations in the catalytic cycle of a highly conserved enzyme.
|
| |
Mol Biochem Parasitol,
170,
93-99.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.A.Borisova,
B.T.Circello,
J.K.Zhang,
W.A.van der Donk,
and
W.W.Metcalf
(2010).
Biosynthesis of rhizocticins, antifungal phosphonate oligopeptides produced by Bacillus subtilis ATCC6633.
|
| |
Chem Biol,
17,
28-37.
|
 |
|
|
|
|
 |
H.Li,
W.Fast,
and
S.J.Benkovic
(2009).
Structural and functional modularity of proteins in the de novo purine biosynthetic pathway.
|
| |
Protein Sci,
18,
881-892.
|
 |
|
|
|
|
 |
W.Zhang,
L.Wang,
Y.Liu,
J.Xu,
G.Zhu,
H.Cang,
X.Li,
M.Bartlam,
K.Hensley,
G.Li,
Z.Rao,
and
X.C.Zhang
(2009).
Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione.
|
| |
Genes Dev,
23,
1387-1392.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.O.Nilsson Lill,
J.Gao,
and
G.L.Waldrop
(2008).
Molecular dynamics simulations of biotin carboxylase.
|
| |
J Phys Chem B,
112,
3149-3156.
|
 |
|
|
|
|
 |
Y.Zhang,
R.H.White,
and
S.E.Ealick
(2008).
Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
|
| |
Biochemistry,
47,
205-217.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Herrera,
R.E.Cahoon,
S.Kumaran,
and
J.Jez
(2007).
Reaction mechanism of glutathione synthetase from Arabidopsis thaliana: site-directed mutagenesis of active site residues.
|
| |
J Biol Chem,
282,
17157-17165.
|
 |
|
|
|
|
 |
P.D.Abeyrathne,
and
J.S.Lam
(2007).
WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core.
|
| |
Mol Microbiol,
65,
1345-1359.
|
 |
|
|
|
|
 |
B.Vergauwen,
D.De Vos,
and
J.J.Van Beeumen
(2006).
Characterization of the bifunctional gamma-glutamate-cysteine ligase/glutathione synthetase (GshF) of Pasteurella multocida.
|
| |
J Biol Chem,
281,
4380-4394.
|
 |
|
|
|
|
 |
L.Masip,
K.Veeravalli,
and
G.Georgiou
(2006).
The many faces of glutathione in bacteria.
|
| |
Antioxid Redox Signal,
8,
753-762.
|
 |
|
|
|
|
 |
M.E.Fraser,
K.Hayakawa,
M.S.Hume,
D.G.Ryan,
and
E.R.Brownie
(2006).
Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase.
|
| |
J Biol Chem,
281,
11058-11065.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Gopal,
I.Borovok,
A.Ofer,
M.Yanku,
G.Cohen,
W.Goebel,
J.Kreft,
and
Y.Aharonowitz
(2005).
A multidomain fusion protein in Listeria monocytogenes catalyzes the two primary activities for glutathione biosynthesis.
|
| |
J Bacteriol,
187,
3839-3847.
|
 |
|
|
|
|
 |
A.Dinescu,
T.R.Cundari,
V.S.Bhansali,
J.L.Luo,
and
M.E.Anderson
(2004).
Function of conserved residues of human glutathione synthetase: implications for the ATP-grasp enzymes.
|
| |
J Biol Chem,
279,
22412-22421.
|
 |
|
|
|
|
 |
J.M.Jez,
and
R.E.Cahoon
(2004).
Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana.
|
| |
J Biol Chem,
279,
42726-42731.
|
 |
|
|
|
|
 |
H.Li,
H.Xu,
D.E.Graham,
and
R.H.White
(2003).
Glutathione synthetase homologs encode alpha-L-glutamate ligases for methanogenic coenzyme F420 and tetrahydrosarcinapterin biosyntheses.
|
| |
Proc Natl Acad Sci U S A,
100,
9785-9790.
|
 |
|
|
|
|
 |
N.Phlippen,
K.Hoffmann,
R.Fischer,
K.Wolf,
and
M.Zimmermann
(2003).
The glutathione synthetase of Schizosaccharomyces pombe is synthesized as a homodimer but retains full activity when present as a heterotetramer.
|
| |
J Biol Chem,
278,
40152-40161.
|
 |
|
|
|
|
 |
J.E.Cronan,
and
G.L.Waldrop
(2002).
Multi-subunit acetyl-CoA carboxylases.
|
| |
Prog Lipid Res,
41,
407-435.
|
 |
|
|
|
|
 |
S.Ramón-Maiques,
A.Marina,
F.Gil-Ortiz,
I.Fita,
and
V.Rubio
(2002).
Structure of acetylglutamate kinase, a key enzyme for arginine biosynthesis and a prototype for the amino acid kinase enzyme family, during catalysis.
|
| |
Structure,
10,
329-342.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.P.Kuzin,
T.Sun,
J.Jorczak-Baillass,
V.L.Healy,
C.T.Walsh,
and
J.R.Knox
(2000).
Enzymes of vancomycin resistance: the structure of D-alanine-D-lactate ligase of naturally resistant Leuconostoc mesenteroides.
|
| |
Structure,
8,
463-470.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
C.Z.Blanchard,
H.M.Holden,
and
G.L.Waldrop
(2000).
Movement of the biotin carboxylase B-domain as a result of ATP binding.
|
| |
J Biol Chem,
275,
16183-16190.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
S.Firestine,
A.Nixon,
S.J.Benkovic,
and
H.M.Holden
(2000).
Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
|
| |
Biochemistry,
39,
8791-8802.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.A.Denessiouk,
and
M.S.Johnson
(2000).
When fold is not important: a common structural framework for adenine and AMP binding in 12 unrelated protein families.
|
| |
Proteins,
38,
310-326.
|
 |
|
|
|
|
 |
C.Li,
T.J.Kappock,
J.Stubbe,
T.M.Weaver,
and
S.E.Ealick
(1999).
X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
|
| |
Structure,
7,
1155-1166.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Polekhina,
P.G.Board,
R.R.Gali,
J.Rossjohn,
and
M.W.Parker
(1999).
Molecular basis of glutathione synthetase deficiency and a rare gene permutation event.
|
| |
EMBO J,
18,
3204-3213.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
F.M.Raushel,
M.M.Benning,
I.Rayment,
and
H.M.Holden
(1999).
The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
8.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
G.Wesenberg,
F.M.Raushel,
and
H.M.Holden
(1999).
Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding.
|
| |
Biochemistry,
38,
2347-2357.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.M.Weaver,
W.Wang,
and
S.E.Ealick
(1999).
Purification, crystallization and preliminary X-ray diffraction data from selenomethionine glycinamide ribonucleotide synthetase.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
518-521.
|
 |
|
|
|
|
 |
F.M.Raushel,
J.B.Thoden,
G.D.Reinhart,
and
H.M.Holden
(1998).
Carbamoyl phosphate synthetase: a crooked path from substrates to products.
|
| |
Curr Opin Chem Biol,
2,
624-632.
|
 |
|
|
|
|
 |
H.M.Holden,
J.B.Thoden,
and
F.M.Raushel
(1998).
Carbamoyl phosphate synthetase: a tunnel runs through it.
|
| |
Curr Opin Struct Biol,
8,
679-685.
|
 |
|
|
|
|
 |
K.A.Denessiouk,
J.V.Lehtonen,
and
M.S.Johnson
(1998).
Enzyme-mononucleotide interactions: three different folds share common structural elements for ATP recognition.
|
| |
Protein Sci,
7,
1768-1771.
|
 |
|
|
|
|
 |
K.A.Denessiouk,
J.V.Lehtonen,
T.Korpela,
and
M.S.Johnson
(1998).
Two "unrelated" families of ATP-dependent enzymes share extensive structural similarities about their cofactor binding sites.
|
| |
Protein Sci,
7,
1136-1146.
|
 |
|
|
|
|
 |
L.Esser,
C.R.Wang,
M.Hosaka,
C.S.Smagula,
T.C.Südhof,
and
J.Deisenhofer
(1998).
Synapsin I is structurally similar to ATP-utilizing enzymes.
|
| |
EMBO J,
17,
977-984.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.McGuire,
K.Huang,
G.Kapadia,
O.Herzberg,
and
D.Dunaway-Mariano
(1998).
Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase.
|
| |
Biochemistry,
37,
13463-13474.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Dideberg,
and
J.Bertrand
(1998).
Tubulin tyrosine ligase: a shared fold with the glutathione synthetase ADP-forming family.
|
| |
Trends Biochem Sci,
23,
57-58.
|
 |
|
|
|
|
 |
T.Nakatsu,
H.Kato,
and
J.Oda
(1998).
Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
|
| |
Nat Struct Biol,
5,
15-19.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.M.Levdikov,
V.V.Barynin,
A.I.Grebenko,
W.R.Melik-Adamyan,
V.S.Lamzin,
and
K.S.Wilson
(1998).
The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
|
| |
Structure,
6,
363-376.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Wang,
T.J.Kappock,
J.Stubbe,
and
S.E.Ealick
(1998).
X-ray crystal structure of glycinamide ribonucleotide synthetase from Escherichia coli.
|
| |
Biochemistry,
37,
15647-15662.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
B.X.Yan,
and
Y.Q.Sun
(1997).
Glycine residues provide flexibility for enzyme active sites.
|
| |
J Biol Chem,
272,
3190-3194.
|
 |
|
|
|
|
 |
C.M.Grant,
F.H.MacIver,
and
I.W.Dawes
(1997).
Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine.
|
| |
Mol Biol Cell,
8,
1699-1707.
|
 |
|
|
|
|
 |
J.A.Bertrand,
G.Auger,
E.Fanchon,
L.Martin,
D.Blanot,
J.van Heijenoort,
and
O.Dideberg
(1997).
Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli.
|
| |
EMBO J,
16,
3416-3425.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
H.M.Holden,
G.Wesenberg,
F.M.Raushel,
and
I.Rayment
(1997).
Structure of carbamoyl phosphate synthetase: a journey of 96 A from substrate to product.
|
| |
Biochemistry,
36,
6305-6316.
|
 |
|
|
|
|
 |
M.Kothe,
B.Eroglu,
H.Mazza,
H.Samudera,
and
S.Powers-Lee
(1997).
Novel mechanism for carbamoyl-phosphate synthetase: a nucleotide switch for functionally equivalent domains.
|
| |
Proc Natl Acad Sci U S A,
94,
12348-12353.
|
 |
|
|
|
|
 |
M.Y.Galperin,
and
E.V.Koonin
(1997).
A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity.
|
| |
Protein Sci,
6,
2639-2643.
|
 |
|
|
|
|
 |
N.Kobayashi,
and
N.Go
(1997).
ATP binding proteins with different folds share a common ATP-binding structural motif.
|
| |
Nat Struct Biol,
4,
6-7.
|
 |
|
|
|
|
 |
D.V.Lueder,
and
M.A.Phillips
(1996).
Characterization of Trypanosoma brucei gamma-glutamylcysteine synthetase, an essential enzyme in the biosynthesis of trypanothione (diglutathionylspermidine).
|
| |
J Biol Chem,
271,
17485-17490.
|
 |
|
|
|
|
 |
J.Cervera,
E.Bendala,
H.G.Britton,
J.Bueso,
Z.Nassif,
C.J.Lusty,
and
V.Rubio
(1996).
Photoaffinity labeling with UMP of lysine 992 of carbamyl phosphate synthetase from Escherichia coli allows identification of the binding site for the pyrimidine inhibitor.
|
| |
Biochemistry,
35,
7247-7255.
|
 |
|
|
|
|
 |
M.McGuire,
L.J.Carroll,
L.Yankie,
S.H.Thrall,
D.Dunaway-Mariano,
O.Herzberg,
B.Jayaram,
and
B.H.Haley
(1996).
Determination of the nucleotide binding site within Clostridium symbiosum pyruvate phosphate dikinase by photoaffinity labeling, site-directed mutagenesis, and structural analysis.
|
| |
Biochemistry,
35,
8544-8552.
|
 |
|
|
|
|
 |
M.R.Redinbo,
S.M.Eide,
R.L.Stone,
J.E.Dixon,
and
T.O.Yeates
(1996).
Crystallization and preliminary structural analysis of Bacillus subtilis adenylosuccinate lyase, an enzyme implicated in infantile autism.
|
| |
Protein Sci,
5,
786-788.
|
 |
|
|
|
|
 |
O.Herzberg,
C.C.Chen,
G.Kapadia,
M.McGuire,
L.J.Carroll,
S.J.Noh,
and
D.Dunaway-Mariano
(1996).
Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
|
| |
Proc Natl Acad Sci U S A,
93,
2652-2657.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.J.Artymiuk,
A.R.Poirrette,
D.W.Rice,
and
P.Willett
(1996).
Biotin carboxylase comes into the fold.
|
| |
Nat Struct Biol,
3,
128-132.
|
 |
|
|
|
|
 |
P.Ullmann,
L.Gondet,
S.Potier,
and
T.J.Bach
(1996).
Cloning of Arabidopsis thaliana glutathione synthetase (GSH2) by functional complementation of a yeast gsh2 mutant.
|
| |
Eur J Biochem,
236,
662-669.
|
 |
|
|
|
|
 |
T.Hara,
H.Kato,
Y.Katsube,
and
J.Oda
(1996).
A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
|
| |
Biochemistry,
35,
11967-11974.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hibi,
T.Nishioka,
H.Kato,
K.Tanizawa,
T.Fukui,
Y.Katsube,
and
J.Oda
(1996).
Structure of the multifunctional loops in the nonclassical ATP-binding fold of glutathione synthetase.
|
| |
Nat Struct Biol,
3,
16-18.
|
 |
|
|
|
|
 |
Y.Inoue,
K.Sugiyama,
H.Ueminami,
S.Izawa,
and
A.Kimura
(1996).
Modification of Escherichia coli B glutathione synthetase with polyethylene glycol for clinical application to enzyme replacement therapy for glutathione deficiency.
|
| |
Clin Diagn Lab Immunol,
3,
663-668.
|
 |
|
|
|
|
 |
C.Fan,
P.C.Moews,
Y.Shi,
C.T.Walsh,
and
J.R.Knox
(1995).
A common fold for peptide synthetases cleaving ATP to ADP: glutathione synthetase and D-alanine:d-alanine ligase of Escherichia coli.
|
| |
Proc Natl Acad Sci U S A,
92,
1172-1176.
|
 |
|
|
|
|
 |
E.Alonso,
and
V.Rubio
(1995).
Affinity cleavage of carbamoyl-phosphate synthetase I localizes regions of the enzyme interacting with the molecule of ATP that phosphorylates carbamate.
|
| |
Eur J Biochem,
229,
377-384.
|
 |
|
|
|
|
 |
J.M.Bollinger,
D.S.Kwon,
G.W.Huisman,
R.Kolter,
and
C.T.Walsh
(1995).
Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase.
|
| |
J Biol Chem,
270,
14031-14041.
|
 |
|
|
|
|
 |
M.Lo Bello,
A.Battistoni,
A.P.Mazzetti,
P.G.Board,
M.Muramatsu,
G.Federici,
and
G.Ricci
(1995).
Site-directed mutagenesis of human glutathione transferase P1-1. Spectral, kinetic, and structural properties of Cys-47 and Lys-54 mutants.
|
| |
J Biol Chem,
270,
1249-1253.
|
 |
|
|
|
|
 |
D.Bossemeyer
(1994).
The glycine-rich sequence of protein kinases: a multifunctional element.
|
| |
Trends Biochem Sci,
19,
201-205.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

