 |
PDBsum entry 1a48

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Atp binding protein
|
PDB id
|
|

|

|
1a48
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.6
- phosphoribosylaminoimidazolesuccinocarboxamide synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate + L-aspartate + ATP = (2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate + ADP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
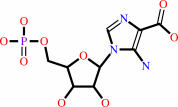 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate
|
+
|
 L-aspartate
L-aspartate
|
+
|
 ATP
ATP
|
=
|
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
6:363-376
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
|
|
V.M.Levdikov,
V.V.Barynin,
A.I.Grebenko,
W.R.Melik-Adamyan,
V.S.Lamzin,
K.S.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The biosynthesis of key metabolic components is of major interest to
biologists. Studies of de novo purine synthesis are aimed at obtaining a deeper
understanding of this central pathway and the development of effective
chemotherapeutic agents. Phosphoribosylaminoimidazolesuccinocarboxamide (SAICAR)
synthase catalyses the seventh step out of ten in the biosynthesis of purine
nucleotides. To date, only one structure of an enzyme involved in purine
biosynthesis has been reported: adenylosuccinate synthetase, which catalyses the
first committed step in the synthesis of AMP from IMP. RESULTS: We report the
first three-dimensional structure of a SAICAR synthase, from Saccharomyces
cerevisiae. It is a monomer with three domains. The first two domains consist of
antiparallel beta sheets and the third is composed of two alpha helices. There
is a long deep cleft made up of residues from all three domains. Comparison of
SAICAR synthases by alignment of their sequences reveals a number of conserved
residues, mostly located in the cleft. The presence of two sulphate ions bound
in the cleft, the structure of SAICAR synthase in complex with ATP and a
comparison of this structure with that of other ATP-dependent proteins point to
the interdomain cleft as the location of the active site. CONCLUSIONS: The
topology of the first domain of SAICAR synthase resembles that of the N-terminal
domain of proteins belonging to the cyclic AMP-dependent protein kinase family.
The fold of the second domain is similar to that of members of the
D-alanine:D-alanine ligase family. Together these enzymes form a new superfamily
of mononucleotide-binding domains. There appears to be no other enzyme, however,
which is composed of the same combination of three domains, with the individual
topologies found in SAICAR synthase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. Schematic representation of the biosynthesis of
purine nucleotides in S. cerevisiae. For each step, the name of
the enzyme and the structural gene (in parentheses) are given.
The reaction catalysed by SAICAR synthase is shown in red.
Abbreviations: AICAR, phosphoribosylaminoimidazolecarboxamide;
AIR, phosphoribosylaminoimidazole; CAIR,
phosphoribosylcarboxyaminoimidazole; FAICAR,
phosphoribosylformylaminoimidazolecarboxamide; FGAM,
phosphoribosylformylglycinamidine; FGAR,
phosphoribosylformylglycinamide; GAR, phosphoribosylglycinamide;
PR, 5-phosphoribosyl; PRA, phosphoribosylamine; PRPP,
5-phosphoribosyl 1-pyrophosphate.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1998,
6,
363-376)
copyright 1998.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
M.Morar,
R.Anand,
A.A.Hoskins,
J.Stubbe,
and
S.E.Ealick
(2006).
Complexed structures of formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima describe a novel ATP binding protein superfamily.
|
| |
Biochemistry,
45,
14880-14895.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.D.Ginder,
D.J.Binkowski,
H.J.Fromm,
and
R.B.Honzatko
(2006).
Nucleotide complexes of Escherichia coli phosphoribosylaminoimidazole succinocarboxamide synthetase.
|
| |
J Biol Chem,
281,
20680-20688.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.H.Zwart,
G.G.Langer,
and
V.S.Lamzin
(2004).
Modelling bound ligands in protein crystal structures.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2230-2239.
|
 |
|
|
|
|
 |
K.Matsuda,
T.Nishioka,
K.Kinoshita,
T.Kawabata,
and
N.Go
(2003).
Finding evolutionary relations beyond superfamilies: fold-based superfamilies.
|
| |
Protein Sci,
12,
2239-2251.
|
 |
|
|
|
|
 |
H.Yamaguchi,
M.Matsushita,
A.C.Nairn,
and
J.Kuriyan
(2001).
Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
|
| |
Mol Cell,
7,
1047-1057.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.J.Kappock,
S.E.Ealick,
and
J.Stubbe
(2000).
Modular evolution of the purine biosynthetic pathway.
|
| |
Curr Opin Chem Biol,
4,
567-572.
|
 |
|
|
|
|
 |
T.Zhou,
M.Daugherty,
N.V.Grishin,
A.L.Osterman,
and
H.Zhang
(2000).
Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily.
|
| |
Structure,
8,
1247-1257.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Li,
T.J.Kappock,
J.Stubbe,
T.M.Weaver,
and
S.E.Ealick
(1999).
X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
|
| |
Structure,
7,
1155-1166.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Roy
(1999).
Multifunctional enzymes and evolution of biosynthetic pathways: retro-evolution by jumps.
|
| |
Proteins,
37,
303-309.
|
 |
|
|
|
|
 |
W.Wang,
T.J.Kappock,
J.Stubbe,
and
S.E.Ealick
(1998).
X-ray crystal structure of glycinamide ribonucleotide synthetase from Escherichia coli.
|
| |
Biochemistry,
37,
15647-15662.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

