 |
PDBsum entry 1fvg

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1fvg
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.4.11
- peptide-methionine (S)-S-oxide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-methionyl-[protein] + [thioredoxin]-disulfide + H2O = L-methionyl- (S)-S-oxide-[protein] + [thioredoxin]-dithiol
|
|
2.
|
[thioredoxin]-disulfide + L-methionine + H2O = L-methionine (S)-S- oxide + [thioredoxin]-dithiol
|
|
 |
 |
 |
 |
 |
L-methionyl-[protein]
|
+
|
[thioredoxin]-disulfide
|
+
|
H2O
|
=
|
L-methionyl- (S)-S-oxide-[protein]
|
+
|
[thioredoxin]-dithiol
|
|
 |
 |
 |
 |
 |
[thioredoxin]-disulfide
Bound ligand (Het Group name = )
matches with 41.67% similarity
|
+
|
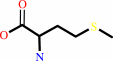 L-methionine
L-methionine
|
+
|
H2O
|
=
|
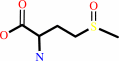 L-methionine (S)-S- oxide
L-methionine (S)-S- oxide
|
+
|
[thioredoxin]-dithiol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
39:13307-13312
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and mechanism of peptide methionine sulfoxide reductase, an "anti-oxidation" enzyme.
|
|
W.T.Lowther,
N.Brot,
H.Weissbach,
B.W.Matthews.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Peptide methionine sulfoxide reductase (MsrA) reverses oxidative damage to both
free methionine and methionine within proteins. As such, it helps protect the
host organism against stochastic damage that can contribute to cell death. The
structure of bovine MsrA has been determined in two different modifications,
both of which provide different insights into the biology of the protein. There
are three cysteine residues located in the vicinity of the active site.
Conformational changes in a glycine-rich C-terminal tail appear to allow all
three thiols to come together and to participate in catalysis. The structures
support a unique, thiol-disulfide exchange mechanism that relies upon an
essential cysteine as a nucleophile and additional conserved residues that
interact with the oxygen atom of the sulfoxide moiety.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.J.Kim,
B.C.Lee,
J.Jeong,
K.J.Lee,
K.Y.Hwang,
V.N.Gladyshev,
and
H.Y.Kim
(2011).
Tandem use of selenocysteine: adaptation of a selenoprotein glutaredoxin for reduction of selenoprotein methionine sulfoxide reductase.
|
| |
Mol Microbiol,
79,
1194-1203.
|
 |
|
|
|
|
 |
H.Y.Kim,
Y.Zhang,
B.C.Lee,
J.R.Kim,
and
V.N.Gladyshev
(2009).
The selenoproteome of Clostridium sp. OhILAs: characterization of anaerobic bacterial selenoprotein methionine sulfoxide reductase A.
|
| |
Proteins,
74,
1008-1017.
|
 |
|
|
|
|
 |
Y.K.Kim,
Y.J.Shin,
W.H.Lee,
H.Y.Kim,
and
K.Y.Hwang
(2009).
Structural and kinetic analysis of an MsrA-MsrB fusion protein from Streptococcus pneumoniae.
|
| |
Mol Microbiol,
72,
699-709.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.R.Salsbury,
S.T.Knutson,
L.B.Poole,
and
J.S.Fetrow
(2008).
Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid.
|
| |
Protein Sci,
17,
299-312.
|
 |
|
|
|
|
 |
K.U.Schallreuter,
K.Rübsam,
N.C.Gibbons,
D.J.Maitland,
B.Chavan,
C.Zothner,
H.Rokos,
and
J.M.Wood
(2008).
Methionine sulfoxide reductases A and B are deactivated by hydrogen peroxide (H2O2) in the epidermis of patients with vitiligo.
|
| |
J Invest Dermatol,
128,
808-815.
|
 |
|
|
|
|
 |
X.H.Zhang,
and
H.Weissbach
(2008).
Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases.
|
| |
Biol Rev Camb Philos Soc,
83,
249-257.
|
 |
|
|
|
|
 |
A.Gand,
M.Antoine,
S.Boschi-Muller,
and
G.Branlant
(2007).
Characterization of the amino acids involved in substrate specificity of methionine sulfoxide reductase A.
|
| |
J Biol Chem,
282,
20484-20491.
|
 |
|
|
|
|
 |
N.Rouhier,
B.Kauffmann,
F.Tete-Favier,
P.Palladino,
P.Gans,
G.Branlant,
J.P.Jacquot,
and
S.Boschi-Muller
(2007).
Functional and structural aspects of poplar cytosolic and plastidial type a methionine sulfoxide reductases.
|
| |
J Biol Chem,
282,
3367-3378.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Gröbe,
M.Reuter,
T.Gursinsky,
B.Söhling,
and
J.R.Andreesen
(2007).
Peroxidase activity of selenoprotein GrdB of glycine reductase and stabilisation of its integrity by components of proprotein GrdE from Eubacterium acidaminophilum.
|
| |
Arch Microbiol,
187,
29-43.
|
 |
|
|
|
|
 |
Z.Lin,
L.C.Johnson,
H.Weissbach,
N.Brot,
M.O.Lively,
and
W.T.Lowther
(2007).
Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function.
|
| |
Proc Natl Acad Sci U S A,
104,
9597-9602.
|
 |
|
|
|
|
 |
D.Sagher,
D.Brunell,
J.F.Hejtmancik,
M.Kantorow,
N.Brot,
and
H.Weissbach
(2006).
Thionein can serve as a reducing agent for the methionine sulfoxide reductases.
|
| |
Proc Natl Acad Sci U S A,
103,
8656-8661.
|
 |
|
|
|
|
 |
D.Sagher,
D.Brunell,
N.Brot,
B.L.Vallee,
and
H.Weissbach
(2006).
Selenocompounds can serve as oxidoreductants with the methionine sulfoxide reductase enzymes.
|
| |
J Biol Chem,
281,
31184-31187.
|
 |
|
|
|
|
 |
H.Y.Kim,
D.E.Fomenko,
Y.E.Yoon,
and
V.N.Gladyshev
(2006).
Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases.
|
| |
Biochemistry,
45,
13697-13704.
|
 |
|
|
|
|
 |
M.Antoine,
A.Gand,
S.Boschi-Muller,
and
G.Branlant
(2006).
Characterization of the amino acids from Neisseria meningitidis MsrA involved in the chemical catalysis of the methionine sulfoxide reduction step.
|
| |
J Biol Chem,
281,
39062-39070.
|
 |
|
|
|
|
 |
N.Brot,
J.F.Collet,
L.C.Johnson,
T.J.Jönsson,
H.Weissbach,
and
W.T.Lowther
(2006).
The thioredoxin domain of Neisseria gonorrhoeae PilB can use electrons from DsbD to reduce downstream methionine sulfoxide reductases.
|
| |
J Biol Chem,
281,
32668-32675.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.J.Meyer,
and
R.Hell
(2005).
Glutathione homeostasis and redox-regulation by sulfhydryl groups.
|
| |
Photosynth Res,
86,
435-457.
|
 |
|
|
|
|
 |
H.Y.Kim,
and
V.N.Gladyshev
(2005).
Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases.
|
| |
PLoS Biol,
3,
e375.
|
 |
|
|
|
|
 |
J.Wu,
F.Neiers,
S.Boschi-Muller,
and
G.Branlant
(2005).
The N-terminal domain of PILB from Neisseria meningitidis is a disulfide reductase that can recycle methionine sulfoxide reductases.
|
| |
J Biol Chem,
280,
12344-12350.
|
 |
|
|
|
|
 |
P.Hu,
E.L.Brodie,
Y.Suzuki,
H.H.McAdams,
and
G.L.Andersen
(2005).
Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus.
|
| |
J Bacteriol,
187,
8437-8449.
|
 |
|
|
|
|
 |
F.Neiers,
A.Kriznik,
S.Boschi-Muller,
and
G.Branlant
(2004).
Evidence for a new sub-class of methionine sulfoxide reductases B with an alternative thioredoxin recognition signature.
|
| |
J Biol Chem,
279,
42462-42468.
|
 |
|
|
|
|
 |
N.Coudevylle,
A.Thureau,
S.Azza,
S.Boshi-Muller,
G.Branlant,
and
M.T.Cung
(2004).
(1)H, (13)C and (15)N resonance assignment of the reduced form of methionine sulfoxide reductase A from Escherichia coli.
|
| |
J Biomol NMR,
30,
363-364.
|
 |
|
|
|
|
 |
A.B.Taylor,
D.M.Benglis,
S.Dhandayuthapani,
and
P.J.Hart
(2003).
Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine.
|
| |
J Bacteriol,
185,
4119-4126.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.V.Smith,
and
J.C.Sacchettini
(2003).
Mycobacterium tuberculosis: a model system for structural genomics.
|
| |
Curr Opin Struct Biol,
13,
658-664.
|
 |
|
|
|
|
 |
M.A.Wilson,
J.L.Collins,
Y.Hod,
D.Ringe,
and
G.A.Petsko
(2003).
The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease.
|
| |
Proc Natl Acad Sci U S A,
100,
9256-9261.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Antoine,
S.Boschi-Muller,
and
G.Branlant
(2003).
Kinetic characterization of the chemical steps involved in the catalytic mechanism of methionine sulfoxide reductase A from Neisseria meningitidis.
|
| |
J Biol Chem,
278,
45352-45357.
|
 |
|
|
|
|
 |
R.Ortenberg,
and
J.Beckwith
(2003).
Functions of thiol-disulfide oxidoreductases in E. coli: redox myths, realities, and practicalities.
|
| |
Antioxid Redox Signal,
5,
403-411.
|
 |
|
|
|
|
 |
A.Olry,
S.Boschi-Muller,
M.Marraud,
S.Sanglier-Cianferani,
A.Van Dorsselear,
and
G.Branlant
(2002).
Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis.
|
| |
J Biol Chem,
277,
12016-12022.
|
 |
|
|
|
|
 |
B.Kauffmann,
F.Favier,
A.Olry,
S.Boschi-Muller,
P.Carpentier,
G.Branlant,
and
A.Aubry
(2002).
Crystallization and preliminary X-ray diffraction studies of the peptide methionine sulfoxide reductase B domain of Neisseria meningitidis PILB.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1467-1469.
|
 |
|
|
|
|
 |
G.V.Kryukov,
R.A.Kumar,
A.Koc,
Z.Sun,
and
V.N.Gladyshev
(2002).
Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase.
|
| |
Proc Natl Acad Sci U S A,
99,
4245-4250.
|
 |
|
|
|
|
 |
H.Ruan,
X.D.Tang,
M.L.Chen,
M.L.Joiner,
G.Sun,
N.Brot,
H.Weissbach,
S.H.Heinemann,
L.Iverson,
C.F.Wu,
T.Hoshi,
M.L.Chen,
M.A.Joiner,
and
S.H.Heinemann
(2002).
High-quality life extension by the enzyme peptide methionine sulfoxide reductase.
|
| |
Proc Natl Acad Sci U S A,
99,
2748-2753.
|
 |
|
|
|
|
 |
R.A.Kumar,
A.Koc,
R.L.Cerny,
and
V.N.Gladyshev
(2002).
Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase.
|
| |
J Biol Chem,
277,
37527-37535.
|
 |
|
|
|
|
 |
V.N.Gladyshev
(2002).
Thioredoxin and peptide methionine sulfoxide reductase: convergence of similar structure and function in distinct structural folds.
|
| |
Proteins,
46,
149-152.
|
 |
|
|
|
|
 |
W.T.Lowther,
H.Weissbach,
F.Etienne,
N.Brot,
and
B.W.Matthews
(2002).
The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB.
|
| |
Nat Struct Biol,
9,
348-352.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Ritz,
and
J.Beckwith
(2001).
Roles of thiol-redox pathways in bacteria.
|
| |
Annu Rev Microbiol,
55,
21-48.
|
 |
|
|
|
|
 |
G.St John,
N.Brot,
J.Ruan,
H.Erdjument-Bromage,
P.Tempst,
H.Weissbach,
and
C.Nathan
(2001).
Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates.
|
| |
Proc Natl Acad Sci U S A,
98,
9901-9906.
|
 |
|
|
|
|
 |
M.J.Barnett,
R.F.Fisher,
T.Jones,
C.Komp,
A.P.Abola,
F.Barloy-Hubler,
L.Bowser,
D.Capela,
F.Galibert,
J.Gouzy,
M.Gurjal,
A.Hong,
L.Huizar,
R.W.Hyman,
D.Kahn,
M.L.Kahn,
S.Kalman,
D.H.Keating,
C.Palm,
M.C.Peck,
R.Surzycki,
D.H.Wells,
K.C.Yeh,
R.W.Davis,
N.A.Federspiel,
and
S.R.Long
(2001).
Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid.
|
| |
Proc Natl Acad Sci U S A,
98,
9883-9888.
|
 |
|
|
|
|
 |
S.Boschi-Muller,
S.Azza,
and
G.Branlant
(2001).
E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide.
|
| |
Protein Sci,
10,
2272-2279.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

