 |
PDBsum entry 1fmt

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Formyltransferase
|
PDB id
|
|

|

|
1fmt
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.2.9
- methionyl-tRNA formyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Coenzymes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionyl-tRNA(fMet) + (6R)-10-formyltetrahydrofolate = N-formyl-L- methionyl-tRNA(fMet) + (6S)-5,6,7,8-tetrahydrofolate + H+
|
 |
 |
 |
 |
 |
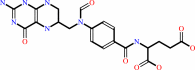 10-formyltetrahydrofolate
10-formyltetrahydrofolate
|
+
|
L-methionyl-tRNA(fMet)
|
=
|
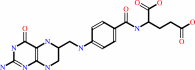 tetrahydrofolate
tetrahydrofolate
|
+
|
N-formylmethionyl-tRNA(fMet)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Embo J
15:4749-4758
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: comparison with glycinamide ribonucleotide formyltransferase.
|
|
E.Schmitt,
S.Blanquet,
Y.Mechulam.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Formylation of the methionyl moiety esterified to the 3' end of tRNA(f)Met is a
key step in the targeting of initiator tRNA towards the translation start
machinery in prokaryotes. Accordingly, the presence of methionyl-tRNA(f)Met
formyltransferase (FMT), the enzyme responsible for this formylation, is
necessary for the normal growth of Escherichia coli. The present work describes
the structure of crystalline E.coli FMT at 2.0 A, resolution. The protein has an
N-terminal domain containing a Rossmann fold. This domain closely resembles that
of the glycinamide ribonucleotide formyltransferase (GARF), an enzyme which,
like FMT, uses N-10 formyltetrahydrofolate as formyl donor. However, FMT can be
distinguished from GARF by a flexible loop inserted within its Rossmann fold. In
addition, FMT possesses a C-terminal domain with a beta-barrel reminiscent of an
OB fold. This latter domain provides a positively charged side oriented towards
the active site. Biochemical evidence is presented for the involvement of these
two idiosyncratic regions (the flexible loop in the N-terminal domain, and the
C-terminal domain) in the binding of the tRNA substrate.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.A.Krupenko
(2009).
FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism.
|
| |
Chem Biol Interact,
178,
84-93.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
J.W.Teo,
P.Thayalan,
D.Beer,
A.S.Yap,
M.Nanjundappa,
X.Ngew,
J.Duraiswamy,
S.Liung,
V.Dartois,
M.Schreiber,
S.Hasan,
M.Cynamon,
N.S.Ryder,
X.Yang,
B.Weidmann,
K.Bracken,
T.Dick,
and
K.Mukherjee
(2006).
Peptide deformylase inhibitors as potent antimycobacterial agents.
|
| |
Antimicrob Agents Chemother,
50,
3665-3673.
|
 |
|
|
|
|
 |
S.N.Reuland,
A.P.Vlasov,
and
S.A.Krupenko
(2006).
Modular organization of FDH: Exploring the basis of hydrolase catalysis.
|
| |
Protein Sci,
15,
1076-1084.
|
 |
|
|
|
|
 |
P.Z.Gatzeva-Topalova,
A.P.May,
and
M.C.Sousa
(2005).
Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance.
|
| |
Biochemistry,
44,
5328-5338.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.D.Breazeale,
A.A.Ribeiro,
A.L.McClerren,
and
C.R.Raetz
(2005).
A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose.
|
| |
J Biol Chem,
280,
14154-14167.
|
 |
|
|
|
|
 |
A.A.Chumanevich,
S.A.Krupenko,
and
C.Davies
(2004).
The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain.
|
| |
J Biol Chem,
279,
14355-14364.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Kessler,
H.Schuhmann,
S.Morneweg,
U.Linne,
and
M.A.Marahiel
(2004).
The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains.
|
| |
J Biol Chem,
279,
7413-7419.
|
 |
|
|
|
|
 |
L.Moulinier,
D.A.Case,
and
T.Simonson
(2003).
Reintroducing electrostatics into protein X-ray structure refinement: bulk solvent treated as a dielectric continuum.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
2094-2103.
|
 |
|
|
|
|
 |
C.Mayer,
and
U.L.RajBhandary
(2002).
Conformational change of Escherichia coli initiator methionyl-tRNA(fMet) upon binding to methionyl-tRNA formyl transferase.
|
| |
Nucleic Acids Res,
30,
2844-2850.
|
 |
|
|
|
|
 |
T.H.Tan,
N.Bochud-Allemann,
E.K.Horn,
and
A.Schneider
(2002).
Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria.
|
| |
Proc Natl Acad Sci U S A,
99,
1152-1157.
|
 |
|
|
|
|
 |
C.Mayer,
A.Stortchevoi,
C.Köhrer,
U.Varshney,
and
U.L.RajBhandary
(2001).
Initiator tRNA and its role in initiation of protein synthesis.
|
| |
Cold Spring Harb Symp Quant Biol,
66,
195-206.
|
 |
|
|
|
|
 |
P.M.Vignais,
B.Billoud,
and
J.Meyer
(2001).
Classification and phylogeny of hydrogenases.
|
| |
FEMS Microbiol Rev,
25,
455-501.
|
 |
|
|
|
|
 |
K.Diederichs,
J.Diez,
G.Greller,
C.Müller,
J.Breed,
C.Schnell,
C.Vonrhein,
W.Boos,
and
W.Welte
(2000).
Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis.
|
| |
EMBO J,
19,
5951-5961.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ibba,
and
D.Soll
(2000).
Aminoacyl-tRNA synthesis.
|
| |
Annu Rev Biochem,
69,
617-650.
|
 |
|
|
|
|
 |
S.Blanquet,
Y.Mechulam,
and
E.Schmitt
(2000).
The many routes of bacterial transfer RNAs after aminoacylation.
|
| |
Curr Opin Struct Biol,
10,
95.
|
 |
|
|
|
|
 |
S.Gite,
Y.Li,
V.Ramesh,
and
U.L.RajBhandary
(2000).
Escherichia coli methionyl-tRNA formyltransferase: role of amino acids conserved in the linker region and in the C-terminal domain on the specific recognition of the initiator tRNA.
|
| |
Biochemistry,
39,
2218-2226.
|
 |
|
|
|
|
 |
D.T.Newton,
M.Niemkiewicz,
R.Y.Lo,
and
D.Mangroo
(1999).
Recognition of the initiator tRNA by the Pseudomonas aeruginosa methionyl-tRNA formyltransferase: importance of the base-base mismatch at the end of the acceptor stem.
|
| |
FEMS Microbiol Lett,
178,
289-298.
|
 |
|
|
|
|
 |
E.Schmitt,
S.Blanquet,
and
Y.Mechulam
(1999).
Crystallization and preliminary X-ray analysis of Escherichia coli methionyl-tRNAMet(f) formyltransferase complexed with formyl-methionyl-tRNAMet(f).
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
332-334.
|
 |
|
|
|
|
 |
M.Fromant,
P.Plateau,
E.Schmitt,
Y.Mechulam,
and
S.Blanquet
(1999).
Receptor site for the 5'-phosphate of elongator tRNAs governs substrate selection by peptidyl-tRNA hydrolase.
|
| |
Biochemistry,
38,
4982-4987.
|
 |
|
|
|
|
 |
S.A.Krupenko,
and
C.Wagner
(1999).
Aspartate 142 is involved in both hydrolase and dehydrogenase catalytic centers of 10-formyltetrahydrofolate dehydrogenase.
|
| |
J Biol Chem,
274,
35777-35784.
|
 |
|
|
|
|
 |
S.Cusack
(1999).
RNA-protein complexes.
|
| |
Curr Opin Struct Biol,
9,
66-73.
|
 |
|
|
|
|
 |
V.Ramesh,
C.Mayer,
M.R.Dyson,
S.Gite,
and
U.L.RajBhandary
(1999).
Induced fit of a peptide loop of methionyl-tRNA formyltransferase triggered by the initiator tRNA substrate.
|
| |
Proc Natl Acad Sci U S A,
96,
875-880.
|
 |
|
|
|
|
 |
E.Schmitt,
M.Panvert,
S.Blanquet,
and
Y.Mechulam
(1998).
Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet.
|
| |
EMBO J,
17,
6819-6826.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Takeuchi,
M.Kawakami,
A.Omori,
T.Ueda,
L.L.Spremulli,
and
K.Watanabe
(1998).
Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure.
|
| |
J Biol Chem,
273,
15085-15090.
|
 |
|
|
|
|
 |
V.Ramesh,
S.Gite,
and
U.L.RajBhandary
(1998).
Functional interaction of an arginine conserved in the sixteen amino acid insertion module of Escherichia coli methionyl-tRNA formyltransferase with determinants for formylation in the initiator tRNA.
|
| |
Biochemistry,
37,
15925-15932.
|
 |
|
|
|
|
 |
V.Ramesh,
S.Gite,
Y.Li,
and
U.L.RajBhandary
(1997).
Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: role of a 16-amino acid insertion module in initiator tRNA recognition.
|
| |
Proc Natl Acad Sci U S A,
94,
13524-13529.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

