
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Electron transport(flavocytochrome)
|
 |
|
Title:
|
 |
The structure of flavocytochromE C sulfide dehydrogenase from a purple phototrophic bacterium chromatium vinosum at 2.5 angstroms resolution
|
|
Structure:
|
 |
FlavocytochromE C sulfide dehydrogenase (flavin-binding subunit). Chain: a, b. Engineered: yes. FlavocytochromE C sulfide dehydrogenase (cytochrome subunit). Chain: c, d. Engineered: yes
|
|
Source:
|
 |
Allochromatium vinosum. Organism_taxid: 1049. Organism_taxid: 1049
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
Z.W.Chen,M.Koh,G.Van Driessche,J.J.Van Beeumen,R.G.Bartsch,T.E.Meyer, M.A.Cusanovich,F.S.Mathews
|
|
Key ref:
|
 |
Z.W.Chen
et al.
(1994).
The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium.
Science,
266,
430-432.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
18-Aug-94
|
Release date:
|
01-Nov-94
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.1.8.2.3
- sulfide-cytochrome-c reductase (flavocytochrome c).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogen sulfide + 2 Fe(III)-[cytochrome c] = sulfur + 2 Fe(II)- [cytochrome c] + H+
|
 |
 |
 |
 |
 |
Hydrogen sulfide
|
+
|
 2
×
ferricytochrome c
2
×
ferricytochrome c
|
=
|
sulfur
|
+
|
 2
×
ferrocytochrome c
2
×
ferrocytochrome c
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Heme c
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
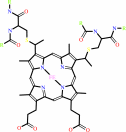
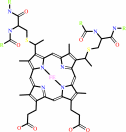
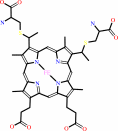 Heme c
Heme c
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
266:430-432
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium.
|
|
Z.W.Chen,
M.Koh,
G.Van Driessche,
J.J.Van Beeumen,
R.G.Bartsch,
T.E.Meyer,
M.A.Cusanovich,
F.S.Mathews.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the heterodimeric flavocytochrome c sulfide dehydrogenase from
Chromatium vinosum was determined at a resolution of 2.53 angstroms. It contains
a glutathione reductase-like flavin-binding subunit and a diheme cytochrome
subunit. The diheme cytochrome folds as two domains, each resembling
mitochondrial cytochrome c, and has an unusual interpropionic acid linkage
joining the two heme groups in the interior of the subunit. The active site of
the flavoprotein subunit contains a catalytically important disulfide bridge
located above the pyrimidine portion of the flavin ring. A tryptophan,
threonine, or tyrosine side chain may provide a partial conduit for electron
transfer to one of the heme groups located 10 angstroms from the flavin.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Marcia,
U.Ermler,
G.Peng,
and
H.Michel
(2010).
A new structure-based classification of sulfide:quinone oxidoreductases.
|
| |
Proteins,
78,
1073-1083.
|
 |
|
|
|
|
 |
D.P.Heuts,
N.S.Scrutton,
W.S.McIntire,
and
M.W.Fraaije
(2009).
What's in a covalent bond? On the role and formation of covalently bound flavin cofactors.
|
| |
FEBS J,
276,
3405-3427.
|
 |
|
|
|
|
 |
J.R.Wallen,
T.C.Mallett,
W.Boles,
D.Parsonage,
C.M.Furdui,
P.A.Karplus,
and
A.Claiborne
(2009).
Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
|
| |
Biochemistry,
48,
9650-9667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.K.Chan,
R.M.Morgan-Kiss,
and
T.E.Hanson
(2009).
Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum.
|
| |
J Bacteriol,
191,
1026-1034.
|
 |
|
|
|
|
 |
M.Marcia,
U.Ermler,
G.Peng,
and
H.Michel
(2009).
The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration.
|
| |
Proc Natl Acad Sci U S A,
106,
9625-9630.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Tomcová,
R.M.Branca,
G.Bodó,
C.Bagyinka,
and
I.K.Smatanová
(2006).
Cross-crystallization method used for the crystallization and preliminary diffraction analysis of a novel di-haem cytochrome c4.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
820-824.
|
 |
|
|
|
|
 |
R.E.Galian,
L.Pastor-Pérez,
M.A.Miranda,
and
J.Pérez-Prieto
(2005).
Intramolecular electron transfer between tyrosine and tryptophan photosensitized by a chiral pi,pi* aromatic ketone.
|
| |
Chemistry,
11,
3443-3448.
|
 |
|
|
|
|
 |
A.Brigé,
D.Leys,
T.E.Meyer,
M.A.Cusanovich,
and
J.J.Van Beeumen
(2002).
The 1.25 A resolution structure of the diheme NapB subunit of soluble nitrate reductase reveals a novel cytochrome c fold with a stacked heme arrangement.
|
| |
Biochemistry,
41,
4827-4836.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.E.Todd,
C.A.Orengo,
and
J.M.Thornton
(2002).
Plasticity of enzyme active sites.
|
| |
Trends Biochem Sci,
27,
419-426.
|
 |
|
|
|
|
 |
V.A.Bamford,
S.Bruno,
T.Rasmussen,
C.Appia-Ayme,
M.R.Cheesman,
B.C.Berks,
and
A.M.Hemmings
(2002).
Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme.
|
| |
EMBO J,
21,
5599-5610.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.W.Chen,
K.Matsushita,
T.Yamashita,
T.A.Fujii,
H.Toyama,
O.Adachi,
H.D.Bellamy,
and
F.S.Mathews
(2002).
Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
|
| |
Structure,
10,
837-849.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Appia-Ayme,
P.J.Little,
Y.Matsumoto,
A.P.Leech,
and
B.C.Berks
(2001).
Cytochrome complex essential for photosynthetic oxidation of both thiosulfate and sulfide in Rhodovulum sulfidophilum.
|
| |
J Bacteriol,
183,
6107-6118.
|
 |
|
|
|
|
 |
D.E.Edmondson,
and
P.Newton-Vinson
(2001).
The covalent FAD of monoamine oxidase: structural and functional role and mechanism of the flavinylation reaction.
|
| |
Antioxid Redox Signal,
3,
789-806.
|
 |
|
|
|
|
 |
K.A.Denessiouk,
V.V.Rantanen,
and
M.S.Johnson
(2001).
Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins.
|
| |
Proteins,
44,
282-291.
|
 |
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
|
|
|
 |
B.M.Hallberg,
T.Bergfors,
K.Bäckbro,
G.Pettersson,
G.Henriksson,
and
C.Divne
(2000).
A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
|
| |
Structure,
8,
79-88.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.J.Studholme,
S.R.Wigneshwereraraj,
M.T.Gallegos,
and
M.Buck
(2000).
Functionality of purified sigma(N) (sigma(54)) and a NifA-like protein from the hyperthermophile Aquifex aeolicus.
|
| |
J Bacteriol,
182,
1616-1623.
|
 |
|
|
|
|
 |
M.Bronstein,
M.Schütz,
G.Hauska,
E.Padan,
and
Y.Shahak
(2000).
Cyanobacterial sulfide-quinone reductase: cloning and heterologous expression.
|
| |
J Bacteriol,
182,
3336-3344.
|
 |
|
|
|
|
 |
O.Vallon
(2000).
New sequence motifs in flavoproteins: evidence for common ancestry and tools to predict structure.
|
| |
Proteins,
38,
95.
|
 |
|
|
|
|
 |
P.N.Mukhopadhyaya,
C.Deb,
C.Lahiri,
and
P.Roy
(2000).
A soxA gene, encoding a diheme cytochrome c, and a sox locus, essential for sulfur oxidation in a new sulfur lithotrophic bacterium.
|
| |
J Bacteriol,
182,
4278-4287.
|
 |
|
|
|
|
 |
V.Kostanjevecki,
A.Brigé,
T.E.Meyer,
M.A.Cusanovich,
Y.Guisez,
and
J.van Beeumen
(2000).
A membrane-bound flavocytochrome c-sulfide dehydrogenase from the purple phototrophic sulfur bacterium Ectothiorhodospira vacuolata.
|
| |
J Bacteriol,
182,
3097-3103.
|
 |
|
|
|
|
 |
A.Mattevi,
G.Tedeschi,
L.Bacchella,
A.Coda,
A.Negri,
and
S.Ronchi
(1999).
Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family.
|
| |
Structure,
7,
745-756.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Trickey,
M.A.Wagner,
M.S.Jorns,
and
F.S.Mathews
(1999).
Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme.
|
| |
Structure,
7,
331-345.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Mewies,
W.S.McIntire,
and
N.S.Scrutton
(1998).
Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs.
|
| |
Protein Sci,
7,
7.
|
 |
|
|
|
|
 |
A.Kadziola,
and
S.Larsen
(1997).
Crystal structure of the dihaem cytochrome c4 from Pseudomonas stutzeri determined at 2.2A resolution.
|
| |
Structure,
5,
203-216.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Devreese,
C.Costa,
H.Demol,
V.Papaefthymiou,
I.Moura,
J.J.Moura,
and
J.Van Beeumen
(1997).
The primary structure of the split-Soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveals an unusual type of diheme cytochrome c.
|
| |
Eur J Biochem,
248,
445-451.
|
 |
|
|
|
|
 |
C.Wodara,
F.Bardischewsky,
and
C.G.Friedrich
(1997).
Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation.
|
| |
J Bacteriol,
179,
5014-5023.
|
 |
|
|
|
|
 |
G.Van Driessche,
M.Koh,
Z.W.Chen,
F.S.Mathews,
T.E.Meyer,
R.G.Bartsch,
M.A.Cusanovich,
and
J.J.Van Beeumen
(1996).
Covalent structure of the flavoprotein subunit of the flavocytochrome c: sulfide dehydrogenase from the purple phototrophic bacterium Chromatium vinosum.
|
| |
Protein Sci,
5,
1753-1764.
|
 |
|
|
|
|
 |
M.T.Bes,
A.L.De Lacey,
M.L.Peleato,
V.M.Fernandez,
and
C.Gómez-Moreno
(1995).
The covalent linkage of a viologen to a flavoprotein reductase transforms it into an oxidase.
|
| |
Eur J Biochem,
233,
593-599.
|
 |
|
|
|
|
 |
U.Ermler,
R.A.Siddiqui,
R.Cramm,
and
B.Friedrich
(1995).
Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution.
|
| |
EMBO J,
14,
6067-6077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Fülöp,
C.J.Ridout,
C.Greenwood,
and
J.Hajdu
(1995).
Crystal structure of the di-haem cytochrome c peroxidase from Pseudomonas aeruginosa.
|
| |
Structure,
3,
1225-1233.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.B.Curry,
M.D.Grabe,
I.V.Kurnikov,
S.S.Skourtis,
D.N.Beratan,
J.J.Regan,
A.J.Aquino,
P.Beroza,
and
J.N.Onuchic
(1995).
Pathways, pathway tubes, pathway docking, and propagators in electron transfer proteins.
|
| |
J Bioenerg Biomembr,
27,
285-293.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

