 |
PDBsum entry 1d7c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1d7c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.99.18
- cellobiose dehydrogenase (acceptor).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-cellobiose + A = D-cellobiono-1,5-lactone + AH2
|
 |
 |
 |
 |
 |
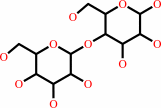 Cellobiose
Cellobiose
|
+
|
 acceptor
acceptor
|
=
|
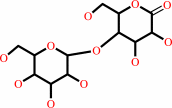 cellobiono-1,5-lactone
cellobiono-1,5-lactone
|
+
|
reduced acceptor
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD; Heme
|
 |
 |
 |
 |
 |
 FAD
FAD
|
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
8:79-88
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
|
|
B.M.Hallberg,
T.Bergfors,
K.Bäckbro,
G.Pettersson,
G.Henriksson,
C.Divne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The fungal oxidoreductase cellobiose dehydrogenase (CDH) degrades
both lignin and cellulose, and is the only known extracellular flavocytochrome.
This haemoflavoenzyme has a multidomain organisation with a b-type cytochrome
domain linked to a large flavodehydrogenase domain. The two domains can be
separated proteolytically to yield a functional cytochrome and a
flavodehydrogenase. Here, we report the crystal structure of the cytochrome
domain of CDH. RESULTS: The crystal structure of the b-type cytochrome domain of
CDH from the wood-degrading fungus Phanerochaete chrysosporium has been
determined at 1.9 A resolution using multiple isomorphous replacement including
anomalous scattering information. Three models of the cytochrome have been
refined: the in vitro prepared cytochrome in its redox-inactive state (pH 7.5)
and redox-active state (pH 4.6), as well as the naturally occurring cytochrome
fragment. CONCLUSIONS: The 190-residue long cytochrome domain of CDH folds as a
beta sandwich with the topology of the antibody Fab V(H) domain. The haem iron
is ligated by Met65 and His163, which confirms previous results from
spectroscopic studies. This is only the second example of a b-type cytochrome
with this ligation, the first being cytochrome b(562). The haem-propionate
groups are surface exposed and, therefore, might play a role in the association
between the cytochrome and flavoprotein domain, and in interdomain electron
transfer. There are no large differences in overall structure of the cytochrome
at redox-active pH as compared with the inactive form, which excludes the
possibility that pH-dependent redox inactivation results from partial
denaturation. From the electron-density map of the naturally occurring
cytochrome, we conclude that it corresponds to the proteolytically prepared
cytochrome domain.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Figure 5. The water-accessible surface of CYT[cdh]. The
surface of the protein molecule is shown in green and the haem
group is in red. The picture was created with INSIGHT II (MSI).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2000,
8,
79-88)
copyright 2000.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.G.Vasilchenko,
K.N.Karapetyan,
O.P.Yershevich,
R.Ludwig,
M.Zamocky,
C.K.Peterbauer,
D.Haltrich,
and
M.L.Rabinovich
(2011).
Cellobiose dehydrogenase of Chaetomium sp. INBI 2-26(-): Structural basis of enhanced activity toward glucose at neutral pH.
|
| |
Biotechnol J,
6,
538-553.
|
 |
|
|
|
|
 |
R.Zhang,
Z.Fan,
and
T.Kasuga
(2011).
Expression of cellobiose dehydrogenase from Neurospora crassa in Pichia pastoris and its purification and characterization.
|
| |
Protein Expr Purif,
75,
63-69.
|
 |
|
|
|
|
 |
A.Kahraman,
R.J.Morris,
R.A.Laskowski,
A.D.Favia,
and
J.M.Thornton
(2010).
On the diversity of physicochemical environments experienced by identical ligands in binding pockets of unrelated proteins.
|
| |
Proteins,
78,
1120-1136.
|
 |
|
|
|
|
 |
L.J.Smith,
A.Kahraman,
and
J.M.Thornton
(2010).
Heme proteins--diversity in structural characteristics, function, and folding.
|
| |
Proteins,
78,
2349-2368.
|
 |
|
|
|
|
 |
R.Ludwig,
W.Harreither,
F.Tasca,
and
L.Gorton
(2010).
Cellobiose dehydrogenase: a versatile catalyst for electrochemical applications.
|
| |
Chemphyschem,
11,
2674-2697.
|
 |
|
|
|
|
 |
S.Pricelius,
R.Ludwig,
N.Lant,
D.Haltrich,
and
G.M.Guebitz
(2009).
Substrate specificity of Myriococcum thermophilum cellobiose dehydrogenase on mono-, oligo-, and polysaccharides related to in situ production of H2O2.
|
| |
Appl Microbiol Biotechnol,
85,
75-83.
|
 |
|
|
|
|
 |
K.H.Sharp,
S.Schneider,
A.Cockayne,
and
M.Paoli
(2007).
Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus.
|
| |
J Biol Chem,
282,
10625-10631.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.P.Kloer,
C.Hagel,
J.Heider,
and
G.E.Schulz
(2006).
Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum.
|
| |
Structure,
14,
1377-1388.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.C.Terwilliger,
H.Klei,
P.D.Adams,
N.W.Moriarty,
and
J.D.Cohn
(2006).
Automated ligand fitting by core-fragment fitting and extension into density.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
915-922.
|
 |
|
|
|
|
 |
K.Igarashi,
M.Yoshida,
H.Matsumura,
N.Nakamura,
H.Ohno,
M.Samejima,
and
T.Nishino
(2005).
Electron transfer chain reaction of the extracellular flavocytochrome cellobiose dehydrogenase from the basidiomycete Phanerochaete chrysosporium.
|
| |
FEBS J,
272,
2869-2877.
|
 |
|
|
|
|
 |
L.Stoica,
N.Dimcheva,
D.Haltrich,
T.Ruzgas,
and
L.Gorton
(2005).
Electrochemical investigation of cellobiose dehydrogenase from new fungal sources on Au electrodes.
|
| |
Biosens Bioelectron,
20,
2010-2018.
|
 |
|
|
|
|
 |
M.Yoshida,
K.Igarashi,
M.Wada,
S.Kaneko,
N.Suzuki,
H.Matsumura,
N.Nakamura,
H.Ohno,
and
M.Samejima
(2005).
Characterization of carbohydrate-binding cytochrome b562 from the white-rot fungus Phanerochaete chrysosporium.
|
| |
Appl Environ Microbiol,
71,
4548-4555.
|
 |
|
|
|
|
 |
C.G.Cheong,
D.W.Wolan,
S.E.Greasley,
P.A.Horton,
G.P.Beardsley,
and
I.A.Wilson
(2004).
Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates.
|
| |
J Biol Chem,
279,
18034-18045.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Kajisa,
M.Yoshida,
K.Igarashi,
A.Katayama,
T.Nishino,
and
M.Samejima
(2004).
Characterization and molecular cloning of cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana.
|
| |
J Biosci Bioeng,
98,
57-63.
|
 |
|
|
|
|
 |
F.A.Rotsaert,
B.M.Hallberg,
S.de Vries,
P.Moenne-Loccoz,
C.Divne,
V.Renganathan,
and
M.H.Gold
(2003).
Biophysical and structural analysis of a novel heme B iron ligation in the flavocytochrome cellobiose dehydrogenase.
|
| |
J Biol Chem,
278,
33224-33231.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.W.Allen,
O.Daltrop,
J.M.Stevens,
and
S.J.Ferguson
(2003).
C-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems.
|
| |
Philos Trans R Soc Lond B Biol Sci,
358,
255-266.
|
 |
|
|
|
|
 |
M.Paoli,
J.Marles-Wright,
and
A.Smith
(2002).
Structure-function relationships in heme-proteins.
|
| |
DNA Cell Biol,
21,
271-280.
|
 |
|
|
|
|
 |
M.Yoshida,
T.Ohira,
K.Igarashi,
H.Nagasawa,
K.Aida,
B.M.Hallberg,
C.Divne,
T.Nishino,
and
M.Samejima
(2001).
Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris.
|
| |
Biosci Biotechnol Biochem,
65,
2050-2057.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

