 |
PDBsum entry 1f6y

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.258
- 5-methyltetrahydrofolate:corrinoid/iron-sulfur protein
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
methyl-Co(III)-[corrinoid Fe-S protein] + (6S)-5,6,7,8-tetrahydrofolate = Co(I)-[corrinoid Fe-S protein] + (6S)-5-methyl-5,6,7,8-tetrahydrofolate + H+
|
 |
 |
 |
 |
 |
[methyl-Co(III) corrinoid Fe-S protein]
|
+
|
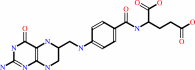 tetrahydrofolate
tetrahydrofolate
|
=
|
[Co(I) corrinoid Fe-S protein]
|
+
|
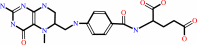 5-methyltetrahydrofolate
5-methyltetrahydrofolate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
8:817-830
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a methyltetrahydrofolate- and corrinoid-dependent methyltransferase.
|
|
T.Doukov,
J.Seravalli,
J.J.Stezowski,
S.W.Ragsdale.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Methyltetrahydrofolate, corrinoid iron-sulfur protein
methyltransferase (MeTr), catalyzes a key step in the Wood-Ljungdahl pathway of
carbon dioxide fixation. It transfers the N5-methyl group from
methyltetrahydrofolate (CH3-H4folate) to a cob(I)amide center in another
protein, the corrinoid iron-sulfur protein. MeTr is a member of a family of
proteins that includes methionine synthase and methanogenic enzymes that
activate the methyl group of methyltetra-hydromethano(or -sarcino)pterin. We
report the first structure of a protein in this family. RESULTS: We determined
the crystal structure of MeTr from Clostridium thermoaceticum at 2.2 A
resolution using multiwavelength anomalous diffraction methods. The overall
architecture presents a new functional class of the versatile triose phosphate
isomerase (TIM) barrel fold. The MeTr tertiary structure is surprisingly similar
to the crystal structures of dihydropteroate synthetases despite sharing less
than 20% sequence identity. This homology permitted the methyl-H4folate binding
site to be modeled. The model suggests extensive conservation of the pterin ring
binding residues in the polar active sites of the methyltransferases and
dihydropteroate synthetases. The most significant structural difference between
these enzymes is in a loop structure above the active site. It is quite open in
MeTr, where it can be modeled as the cobalamin binding site. CONCLUSIONS: The
MeTr structure consists of a TIM barrel that embeds methyl-H4folate and
cobamide. All related methyltransferases are predicted to fold into a similar
TIM barrel pattern and have a similar pterin and cobamide binding site. The
observed structure is consistent with either a 'front' (N5) or 'back' (C8a) side
protonation of CH3-H4folate, a key step that enhances the electrophilic
character of the methyl group, activating it for nucleophilic attack by Co(I).

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 7.
Figure 7. Common structural arrangement for
cobamide-dependent enzymes. (a). The cobalamin coordinates from
the 1bmt structure were used to manually dock the cobalamin
molecule into the TIM barrel cavity by avoiding van der Waals
clashes, but minimizing N5 to Co distance. (b) Interactions
between cobalamin and methylmalonyl-CoA mutase [59 and 60]. The
figures were produced using QUANTA (Molecular Simulations Inc.).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2000,
8,
817-830)
copyright 2000.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Kung,
N.Ando,
T.I.Doukov,
L.C.Blasiak,
G.Bender,
J.Seravalli,
S.W.Ragsdale,
and
C.L.Drennan
(2012).
Visualizing molecular juggling within a B12-dependent methyltransferase complex.
|
| |
Nature,
484,
265-269.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Zhu,
X.Gu,
S.Zhang,
Y.Liu,
Z.X.Huang,
and
X.Tan
(2011).
Efficient expression and purification of methyltransferase in acetyl-coenzyme a synthesis pathway of the human pathogen Clostridium Difficile.
|
| |
Protein Expr Purif,
78,
86-93.
|
 |
|
|
|
|
 |
J.Seravalli,
and
S.W.Ragsdale
(2008).
Pulse-chase studies of the synthesis of acetyl-CoA by carbon monoxide dehydrogenase/acetyl-CoA synthase: evidence for a random mechanism of methyl and carbonyl addition.
|
| |
J Biol Chem,
283,
8384-8394.
|
 |
|
|
|
|
 |
R.G.Matthews,
M.Koutmos,
and
S.Datta
(2008).
Cobalamin-dependent and cobamide-dependent methyltransferases.
|
| |
Curr Opin Struct Biol,
18,
658-666.
|
 |
|
|
|
|
 |
S.W.Ragsdale,
and
E.Pierce
(2008).
Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation.
|
| |
Biochim Biophys Acta,
1784,
1873-1898.
|
 |
|
|
|
|
 |
S.W.Ragsdale
(2008).
Enzymology of the wood-Ljungdahl pathway of acetogenesis.
|
| |
Ann N Y Acad Sci,
1125,
129-136.
|
 |
|
|
|
|
 |
S.W.Ragsdale
(2008).
Catalysis of methyl group transfers involving tetrahydrofolate and B(12).
|
| |
Vitam Horm,
79,
293-324.
|
 |
|
|
|
|
 |
T.A.Stich,
J.Seravalli,
S.Venkateshrao,
T.G.Spiro,
S.W.Ragsdale,
and
T.C.Brunold
(2006).
Spectroscopic studies of the corrinoid/iron-sulfur protein from Moorella thermoacetica.
|
| |
J Am Chem Soc,
128,
5010-5020.
|
 |
|
|
|
|
 |
T.Svetlitchnaia,
V.Svetlitchnyi,
O.Meyer,
and
H.Dobbek
(2006).
Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis.
|
| |
Proc Natl Acad Sci U S A,
103,
14331-14336.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Pejchal,
and
M.L.Ludwig
(2005).
Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication.
|
| |
PLoS Biol,
3,
e31.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.S.Dorweiler,
R.G.Finke,
and
R.G.Matthews
(2003).
Cobalamin-dependent methionine synthase: probing the role of the axial base in catalysis of methyl transfer between methyltetrahydrofolate and exogenous cob(I)alamin or cob(I)inamide.
|
| |
Biochemistry,
42,
14653-14662.
|
 |
|
|
|
|
 |
R.Banerjee,
and
S.W.Ragsdale
(2003).
The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes.
|
| |
Annu Rev Biochem,
72,
209-247.
|
 |
|
|
|
|
 |
B.Hao,
W.Gong,
T.K.Ferguson,
C.M.James,
J.A.Krzycki,
and
M.K.Chan
(2002).
A new UAG-encoded residue in the structure of a methanogen methyltransferase.
|
| |
Science,
296,
1462-1466.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Naidu,
and
S.W.Ragsdale
(2001).
Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica.
|
| |
J Bacteriol,
183,
3276-3281.
|
 |
|
|
|
|
 |
G.Gottschalk,
and
R.K.Thauer
(2001).
The Na(+)-translocating methyltransferase complex from methanogenic archaea.
|
| |
Biochim Biophys Acta,
1505,
28-36.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

