 |
PDBsum entry 1f2d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
1-aminocyclopropane-1-carboxylate deaminase
|
|
Structure:
|
 |
1-aminocyclopropane-1-carboxylate deaminase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Williopsis saturnus. Organism_taxid: 4906. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Octamer (from
)
Octamer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.221
|
R-free:
|
0.268
|
|
|
Authors:
|
 |
M.Yao,T.Ose,H.Sugimoto,A.Horiuchi,A.Nakagawa,D.Yokoi,T.Murakami, M.Honma,S.Wakatsuki,I.Tanaka
|
Key ref:

|
 |
M.Yao
et al.
(2000).
Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J Biol Chem,
275,
34557-34565.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
24-May-00
|
Release date:
|
20-Dec-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q7M523
(1A1D_CYBSA) -
1-aminocyclopropane-1-carboxylate deaminase from Cyberlindnera saturnus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
341 a.a.
341 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.99.7
- 1-aminocyclopropane-1-carboxylate deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-aminocyclopropane-1-carboxylate + H2O = 2-oxobutanoate + NH4+
|
 |
 |
 |
 |
 |
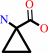 1-aminocyclopropane-1-carboxylate
1-aminocyclopropane-1-carboxylate
|
+
|
H2O
|
=
|
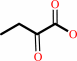 2-oxobutanoate
2-oxobutanoate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
275:34557-34565
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
|
|
M.Yao,
T.Ose,
H.Sugimoto,
A.Horiuchi,
A.Nakagawa,
S.Wakatsuki,
D.Yokoi,
T.Murakami,
M.Honma,
I.Tanaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The pyridoxal 5'-phosphate (PLP)-dependent enzyme
1-aminocyclopropane-1-carboxylate deaminase (ACCD) catalyzes a reaction that
involves a ring opening of cyclopropanoid amino acid, yielding
alpha-ketobutyrate and ammonia. Unlike other PLP-dependent enzymes, this enzyme
has no alpha-hydrogen atom in the substrate. Thus, a unique mechanism for the
bond cleavage is expected. The crystal structure of ACCD from Hansenula saturnus
has been determined at 2.0 A resolution by the multiple wavelength anomalous
diffraction method using mercury atoms as anomalous scatterers. The model was
built on the electron density map, which was obtained by the density averaging
of multiple crystal forms. The final model was refined to an R-factor of 22.5%
and an R(free)-factor of 26.8%. The ACCD folds into two domains, each of which
has an open twisted alpha/beta structure similar to the beta-subunit of
tryptophan synthase. However, in ACCD, unlike in other members of the beta
family of PLP-dependent enzymes, PLP is buried deep in the molecule. The
structure provides the first view of the catalytic center of the cyclopropane
ring opening.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 8.
Fig. 8. Hydrogen bonding networks around the active site.
|
 |
Figure 9.
Fig. 9. A detailed view of the PLP-binding site and
proposed substrate complex. ACC is modeled on the electron
density for the bound sulfoxide ion. Thin bonds are hypothetical
drawings of PLP and the bound substrate (see "Reaction
Mechanism").
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2000,
275,
34557-34565)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Duan,
K.M.Müller,
T.C.Charles,
S.Vesely,
and
B.R.Glick
(2009).
1-Aminocyclopropane-1-Carboxylate (ACC) Deaminase Genes in Rhizobia from Southern Saskatchewan.
|
| |
Microb Ecol,
57,
423-436.
|
 |
|
|
|
|
 |
B.Todorovic,
and
B.R.Glick
(2008).
The interconversion of ACC deaminase and D: -cysteine desulfhydrase by directed mutagenesis.
|
| |
Planta,
229,
193-205.
|
 |
|
|
|
|
 |
C.Prigent-Combaret,
D.Blaha,
J.F.Pothier,
L.Vial,
M.A.Poirier,
F.Wisniewski-Dyé,
and
Y.Moënne-Loccoz
(2008).
Physical organization and phylogenetic analysis of acdR as leucine-responsive regulator of the 1-aminocyclopropane-1-carboxylate deaminase gene acdS in phytobeneficial Azospirillum lipoferum 4B and other Proteobacteria.
|
| |
FEMS Microbiol Ecol,
65,
202-219.
|
 |
|
|
|
|
 |
D.Blaha,
C.Prigent-Combaret,
M.S.Mirza,
and
Y.Moënne-Loccoz
(2006).
Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography.
|
| |
FEMS Microbiol Ecol,
56,
455-470.
|
 |
|
|
|
|
 |
A.Riemenschneider,
R.Wegele,
A.Schmidt,
and
J.Papenbrock
(2005).
Isolation and characterization of a D-cysteine desulfhydrase protein from Arabidopsis thaliana.
|
| |
FEBS J,
272,
1291-1304.
|
 |
|
|
|
|
 |
B.R.Glick
(2005).
Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase.
|
| |
FEMS Microbiol Lett,
251,
1-7.
|
 |
|
|
|
|
 |
K.H.Jhee,
and
W.D.Kruger
(2005).
The role of cystathionine beta-synthase in homocysteine metabolism.
|
| |
Antioxid Redox Signal,
7,
813-822.
|
 |
|
|
|
|
 |
K.Thomazeau,
G.Curien,
R.Dumas,
and
V.Biou
(2001).
Crystal structure of threonine synthase from Arabidopsis thaliana.
|
| |
Protein Sci,
10,
638-648.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Meier,
M.Janosik,
V.Kery,
J.P.Kraus,
and
P.Burkhard
(2001).
Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein.
|
| |
EMBO J,
20,
3910-3916.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

