 |
PDBsum entry 1ebf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ebf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.3
- homoserine dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Threonine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-homoserine + NAD+ = L-aspartate 4-semialdehyde + NADH + H+
|
|
2.
|
L-homoserine + NADP+ = L-aspartate 4-semialdehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
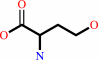 L-homoserine
L-homoserine
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
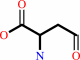 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 L-homoserine
L-homoserine
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
7:238-244
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
|
|
B.DeLaBarre,
P.R.Thompson,
G.D.Wright,
A.M.Berghuis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the antifungal drug target homoserine dehydrogenase (HSD) was
determined from Saccharomyces cerevisiae in apo and holo forms, and as a ternary
complex with bound products, by X-ray diffraction. The three forms show that the
enzyme is a dimer, with each monomer composed of three regions, the
nucleotide-binding region, the dimerization region and the catalytic region. The
dimerization and catalytic regions have novel folds, whereas the fold of the
nucleotide-binding region is a variation on the Rossmann fold. The novel folds
impose a novel composition and arrangement of active site residues when compared
to all other currently known oxidoreductases. This observation, in conjunction
with site-directed mutagenesis of active site residues and steady-state kinetic
measurements, suggest that HSD exhibits a new variation on dehydrogenase
chemistry.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Electron density maps for the two crystal forms of
HSD. a, Stereo view of a 2F[o]- F[c] electron density map
calculated with coefficients from the final tetragonal crystal
form model and contoured at 1  .
The portion of the molecule shown here is the dimer interfacial
region of the extended .
The portion of the molecule shown here is the dimer interfacial
region of the extended  -sheet
and is composed of residues 320 -335 from monomer A, and
residues 332 -335 and 319 -325 from monomer B. b, Stereo view of
the F[o]- F[c] simulated annealing omit map for the NAD^+
molecule in the tetragonal crystal form, contoured at 2 -sheet
and is composed of residues 320 -335 from monomer A, and
residues 332 -335 and 319 -325 from monomer B. b, Stereo view of
the F[o]- F[c] simulated annealing omit map for the NAD^+
molecule in the tetragonal crystal form, contoured at 2  .
c, Stereo view of the F[o]- F[c] simulated annealing omit map
for the NADA and l-Hse molecules in the monoclinic crystal form,
contoured at 2 .
c, Stereo view of the F[o]- F[c] simulated annealing omit map
for the NADA and l-Hse molecules in the monoclinic crystal form,
contoured at 2  . .
|
 |
Figure 5.
Figure 5. Proposed reaction mechanisms of hydride transfer for
HSD. a, Probable reaction mechanism for the forward direction
with the substrate in the aldehyde form. Asp 214, Glu 208 and
Wat460 serve to bind the substrate, whereas Lys 223, oriented by
Asp 219, donates a proton to the C4 oxygen of l-ASA. The
cofactor NADH delivers its pro-S hydride to the C4 carbon of
l-ASA. Arrows indicate electron flow. b, Alternative reaction
mechanism for the forward direction with the substrate in the
gem-diol form. The main difference from the reaction proposed
for the aldehyde form is that Lys 223 donates a proton to the
hydroxyl group, which departs as a water molecule, and Asp 219
hydrogen bonds to the hydroxyl group, which remains in the
product alcohol.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(2000,
7,
238-244)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.J.Sangari,
J.Pérez-Gil,
L.Carretero-Paulet,
J.M.García-Lobo,
and
M.Rodríguez-Concepción
(2010).
A new family of enzymes catalyzing the first committed step of the methylerythritol 4-phosphate (MEP) pathway for isoprenoid biosynthesis in bacteria.
|
| |
Proc Natl Acad Sci U S A,
107,
14081-14086.
|
 |
|
|
|
|
 |
C.C.Lo,
C.A.Bonner,
G.Xie,
M.D'Souza,
and
R.A.Jensen
(2009).
Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways.
|
| |
Microbiol Mol Biol Rev,
73,
594-651.
|
 |
|
|
|
|
 |
E.Di Cera
(2006).
A structural perspective on enzymes activated by monovalent cations.
|
| |
J Biol Chem,
281,
1305-1308.
|
 |
|
|
|
|
 |
L.N.Kinch,
Y.Qi,
T.J.Hubbard,
and
N.V.Grishin
(2003).
CASP5 target classification.
|
| |
Proteins,
53,
340-351.
|
 |
|
|
|
|
 |
S.L.Jacques,
I.A.Mirza,
L.Ejim,
K.Koteva,
D.W.Hughes,
K.Green,
R.Kinach,
J.F.Honek,
H.K.Lai,
A.M.Berghuis,
and
G.D.Wright
(2003).
Enzyme-assisted suicide: molecular basis for the antifungal activity of 5-hydroxy-4-oxonorvaline by potent inhibition of homoserine dehydrogenase.
|
| |
Chem Biol,
10,
989-995.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Ogawa,
and
C.Toyoshima
(2002).
Homology modeling of the cation binding sites of Na+K+-ATPase.
|
| |
Proc Natl Acad Sci U S A,
99,
15977-15982.
|
 |
|
|
|
|
 |
S.L.Jacques,
L.J.Ejim,
and
G.D.Wright
(2001).
Homoserine dehydrogenase from Saccharomyces cerevisiae: kinetic mechanism and stereochemistry of hydride transfer.
|
| |
Biochim Biophys Acta,
1544,
42-54.
|
 |
|
|
|
|
 |
E.Johansson,
J.J.Steffens,
Y.Lindqvist,
and
G.Schneider
(2000).
Crystal structure of saccharopine reductase from Magnaporthe grisea, an enzyme of the alpha-aminoadipate pathway of lysine biosynthesis.
|
| |
Structure,
8,
1037-1047.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

