 |
PDBsum entry 1u1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of e. Coli uridine phosphorylase complexed to 5- benzylacyclouridine (bau)
|
|
Structure:
|
 |
Uridine phosphorylase. Chain: a, b, c, d, e, f. Synonym: urdpase. Upase. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: udp, b3831. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Hexamer (from
)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.219
|
R-free:
|
0.249
|
|
|
Authors:
|
 |
W.Bu,E.C.Settembre,S.E.Ealick
|
Key ref:

|
 |
W.Bu
et al.
(2005).
Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr D Biol Crystallogr,
61,
863-872.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
15-Jul-04
|
Release date:
|
05-Jul-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P12758
(UDP_ECOLI) -
Uridine phosphorylase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
253 a.a.
253 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.3
- uridine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uridine + phosphate = alpha-D-ribose 1-phosphate + uracil
|
 |
 |
 |
 |
 |
uridine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
phosphate
Bound ligand (Het Group name = )
matches with 54.17% similarity
|
=
|
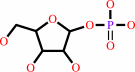 alpha-D-ribose 1-phosphate
alpha-D-ribose 1-phosphate
|
+
|
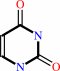 uracil
uracil
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
61:863-872
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
|
|
W.Bu,
E.C.Settembre,
M.H.el Kouni,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Uridine phosphorylase (UP) catalyzes the reversible phosphorolysis of uridine to
uracil and ribose 1-phosphate and is a key enzyme in the pyrimidine-salvage
pathway. Escherichia coli UP is structurally homologous to E. coli purine
nucleoside phosphorylase and other members of the type I family of nucleoside
phosphorylases. The structures of 5-benzylacyclouridine,
5-phenylthioacyclouridine, 5-phenylselenenylacyclouridine, 5-m-benzyloxybenzyl
acyclouridine and 5-m-benzyloxybenzyl barbituric acid acyclonucleoside bound to
the active site of E. coli UP have been determined, with resolutions ranging
from 1.95 to 2.3 A. For all five complexes the acyclo sugar moiety binds to the
active site in a conformation that mimics the ribose ring of the natural
substrates. Surprisingly, the terminal hydroxyl group occupies the position of
the nonessential 5'-hydroxyl substituent of the substrate rather than the
3'-hydroxyl group, which is normally required for catalytic activity. Until
recently, inhibitors of UP were designed with limited structural knowledge of
the active-site residues. These structures explain the basis of inhibition for
this series of acyclouridine analogs and suggest possible additional avenues for
future drug-design efforts. Furthermore, the studies can be extended to design
inhibitors of human UP, for which no X-ray structure is available.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2
Structure of UP shown in ribbon representation. (a) UP monomer with [beta] -strands
in blue and [alpha] -helices in green. BAU and phosphate are shown in stick
representation bound at the active site. C atoms are colored green, N atoms blue, O atoms
red and P atoms pink. (b) UP hexamer shown in ribbon representation with BAU (orange) and
phosphate (red) shown bound at the active sites. Dimers with greater buried surface area
are shown in similar colors. This figure was prepared with MOLSCRIPT (Kraulis, 1991
[Kraulis, P. J. (1991). J. Appl. Cryst. 24, 946-950.]-[bluearr.gif] ) and RASTER3D
(Merritt & Bacon, 1997 [Merritt, E. A. & Bacon, D. J. (1997). Methods Enzymol. 277,
505-524.]-[bluearr.gif] ).
|
 |
Figure 3.
Figure 3
Schematic representation of the UP active site with bound (a) 5-fluorouridine and
phosphate, (b) BAU and phosphate or (c) BBBA. Hydrogen bonds are shown in dashed lines.
The active-site hydrophobic pocket is indicated by a solid line flanked by the
participating residues.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2005,
61,
863-872)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Paul,
S.E.O'Leary,
K.Rajashankar,
W.Bu,
A.Toms,
E.C.Settembre,
J.M.Sanders,
T.P.Begley,
and
S.E.Ealick
(2010).
Glycal formation in crystals of uridine phosphorylase.
|
| |
Biochemistry,
49,
3499-3509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.T.Larson,
D.G.Mudeppa,
J.R.Gillespie,
N.Mueller,
A.J.Napuli,
J.A.Arif,
J.Ross,
T.L.Arakaki,
A.Lauricella,
G.Detitta,
J.Luft,
F.Zucker,
C.L.Verlinde,
E.Fan,
W.C.Van Voorhis,
F.S.Buckner,
P.K.Rathod,
W.G.Hol,
and
E.A.Merritt
(2010).
The crystal structure and activity of a putative trypanosomal nucleoside phosphorylase reveal it to be a homodimeric uridine phosphorylase.
|
| |
J Mol Biol,
396,
1244-1259.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.P.Roosild,
and
S.Castronovo
(2010).
Active site conformational dynamics in human uridine phosphorylase 1.
|
| |
PLoS One,
5,
e12741.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.P.Roosild,
S.Castronovo,
M.Fabbiani,
and
G.Pizzorno
(2009).
Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy.
|
| |
BMC Struct Biol,
9,
14.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

