
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.2.4.2.22
- xanthine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
XMP + diphosphate = xanthine + 5-phospho-alpha-D-ribose 1-diphosphate
|
 |
 |
 |
 |
 |
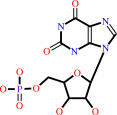 XMP
XMP
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 92.00% similarity
|
=
|
 xanthine
xanthine
|
+
|
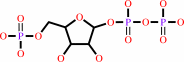 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D:
E.C.2.4.2.8
- hypoxanthine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + diphosphate = hypoxanthine + 5-phospho-alpha-D-ribose 1-diphosphate
|
 |
 |
 |
 |
 |
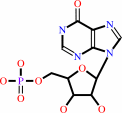 IMP
IMP
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 95.83% similarity
|
=
|
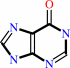 hypoxanthine
hypoxanthine
|
+
|
 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:14485-14494
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of the Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase-GMP and -IMP complexes: comparison of purine binding interactions with the XMP complex.
|
|
A.Héroux,
E.L.White,
L.J.Ross,
D.W.Borhani.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of the guanosine 5'-monophosphate (GMP) and inosine
5'-monophosphate (IMP) complexes of Toxoplasma gondii hypoxanthine-guanine
phosphoribosyltransferase (HGPRT) have been determined at 1.65 and 1.90 A
resolution. These complexes, which crystallize in space groups P2(1) (a = 65.45
A, b = 90.84 A, c = 80. 26 A, and beta = 92.53 degrees ) and P2(1)2(1)2(1) (a =
84.54 A, b = 102.44 A, and c = 108.83 A), each comprise a tetramer in the
crystallographic asymmetric unit. All active sites in the tetramers are fully
occupied by the nucleotide. Comparison of these structures with that of the
xanthosine 5'-monophosphate (XMP)-pyrophosphate-Mg(2+) ternary complex reported
in the following article [Héroux, A., et al. (1999) Biochemistry 38,
14495-14506] shows how T. gondii HGPRT is able to recognize guanine,
hypoxanthine, and xanthine as substrates, and suggests why the human enzyme
cannot use xanthine efficiently. Comparison with the apoenzyme reveals the
structural changes that occur upon binding of purines and ribose 5'-phosphate to
HGPRT. Two structural features important to the HGPRT mechanism, a previously
unrecognized active site loop (loop III', residues 180-184) and an active site
peptide bond (Leu78-Lys79) that adopts both the cis and the trans
configurations, are presented.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Deng,
R.Callender,
V.L.Schramm,
and
C.Grubmeyer
(2010).
Pyrophosphate activation in hypoxanthine--guanine phosphoribosyltransferase with transition state analogue.
|
| |
Biochemistry,
49,
2705-2714.
|
 |
|
|
|
|
 |
P.Gayathri,
I.N.Sujay Subbayya,
C.S.Ashok,
T.S.Selvi,
H.Balaram,
and
M.R.Murthy
(2008).
Crystal structure of a chimera of human and Plasmodium falciparum hypoxanthine guanine phosphoribosyltransferases provides insights into oligomerization.
|
| |
Proteins,
73,
1010-1020.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.S.Monzani,
S.Trapani,
O.H.Thiemann,
and
G.Oliva
(2007).
Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
|
| |
BMC Struct Biol,
7,
59.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Raman,
C.S.Ashok,
S.I.Subbayya,
R.P.Anand,
S.T.Selvi,
and
H.Balaram
(2005).
Plasmodium falciparum hypoxanthine guanine phosphoribosyltransferase. Stability studies on the product-activated enzyme.
|
| |
FEBS J,
272,
1900-1911.
|
 |
|
|
|
|
 |
K.Chaudhary,
R.G.Donald,
M.Nishi,
D.Carter,
B.Ullman,
and
D.S.Roos
(2005).
Differential localization of alternatively spliced hypoxanthine-xanthine-guanine phosphoribosyltransferase isoforms in Toxoplasma gondii.
|
| |
J Biol Chem,
280,
22053-22059.
|
 |
|
|
|
|
 |
J.Duan,
L.Nilsson,
and
B.Lambert
(2004).
Structural and functional analysis of mutations at the human hypoxanthine phosphoribosyl transferase (HPRT1) locus.
|
| |
Hum Mutat,
23,
599-611.
|
 |
|
|
|
|
 |
R.V.Dumitru,
and
S.W.Ragsdale
(2004).
Mechanism of 4-(beta-D-ribofuranosyl)aminobenzene 5'-phosphate synthase, a key enzyme in the methanopterin biosynthetic pathway.
|
| |
J Biol Chem,
279,
39389-39395.
|
 |
|
|
|
|
 |
D.You,
Q.Chen,
Y.Liang,
J.An,
R.Li,
X.Gu,
M.Luo,
and
X.D.Su
(2003).
Protein preparation, crystallization and preliminary X-ray crystallographic studies of a thermostable hypoxanthine-guanine phosphoribosyltransferase from Thermoanaerobacter tengcongensis.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1863-1865.
|
 |
|
|
|
|
 |
C.Bashor,
J.M.Denu,
R.G.Brennan,
and
B.Ullman
(2002).
Kinetic mechanism of adenine phosphoribosyltransferase from Leishmania donovani.
|
| |
Biochemistry,
41,
4020-4031.
|
 |
|
|
|
|
 |
A.M.Aronov,
N.R.Munagala,
I.D.Kuntz,
and
C.C.Wang
(2001).
Virtual screening of combinatorial libraries across a gene family: in search of inhibitors of Giardia lamblia guanine phosphoribosyltransferase.
|
| |
Antimicrob Agents Chemother,
45,
2571-2576.
|
 |
|
|
|
|
 |
N.Munagala,
V.J.Basus,
and
C.C.Wang
(2001).
Role of the flexible loop of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus in enzyme catalysis.
|
| |
Biochemistry,
40,
4303-4311.
|
 |
|
|
|
|
 |
A.Héroux,
E.L.White,
L.J.Ross,
A.P.Kuzin,
and
D.W.Borhani
(2000).
Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis.
|
| |
Structure,
8,
1309-1318.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

