 |
PDBsum entry 1mjt

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1mjt
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.47
- nitric-oxide synthase (flavodoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3 reduced [flavodoxin] + 2 L-arginine + 4 O2 = 3 oxidized [flavodoxin] + 2 L-citrulline + 2 nitric oxide + 4 H2O + 5 H+
|
 |
 |
 |
 |
 |
3
×
reduced [flavodoxin]
Bound ligand (Het Group name = )
matches with 41.18% similarity
|
+
|
 2
×
L-arginine
2
×
L-arginine
|
+
|
 4
×
O2
4
×
O2
|
=
|
3
×
oxidized [flavodoxin]
|
+
|
 2
×
L-citrulline
2
×
L-citrulline
|
+
|
 2
×
nitric oxide
2
×
nitric oxide
|
+
|
4
×
H2O
|
+
|
5
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
5,6,7,8-tetrahydrobiopterin; Ferriheme b
|
 |
 |
 |
 |
 |
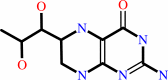 5,6,7,8-tetrahydrobiopterin
5,6,7,8-tetrahydrobiopterin
|
Ferriheme b
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
10:1687-1696
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of SANOS, a bacterial nitric oxide synthase oxygenase protein from Staphylococcus aureus.
|
|
L.E.Bird,
J.Ren,
J.Zhang,
N.Foxwell,
A.R.Hawkins,
I.G.Charles,
D.K.Stammers.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prokaryotic genes related to the oxygenase domain of mammalian nitric oxide
synthases (NOSs) have recently been identified. Although they catalyze the same
reaction as the eukaryotic NOS oxygenase domain, their biological function(s)
are unknown. In order to explore rationally the biochemistry and evolution of
the prokaryotic NOS family, we have determined the crystal structure of SANOS,
from methicillin-resistant Staphylococcus aureus (MRSA), to 2.4 A. Haem and
S-ethylisothiourea (SEITU) are bound at the SANOS active site, while the
intersubunit site, occupied by the redox cofactor tetrahydrobiopterin (H(4)B) in
mammalian NOSs, has NAD(+) bound in SANOS. In common with all bacterial NOSs,
SANOS lacks the N-terminal extension responsible for stable dimerization in
mammalian isoforms, but has alternative interactions to promote dimer formation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. Detailed Structural Analysis of SANOS(A) Stereo
diagram showing the dimer interface of SANOS. The main chains
are shown as ribbons and coils, with the A chain colored green
and the B chain colored blue. The side chains of key residues
involved in the interface interactions are shown as balls and
sticks and colored orange and cyan for the A and B chains,
respectively. The yellow dashed lines represent the hydrogen
bonds between the two chains. The four segments from each chain
are labeled I-IV (residues 233-240, 259-280, 288-291, and
314-330, respectively).(B) Electrostatic surface (A chain) and
ribbons (B chain) showing the charge distribution on the
molecular surface and the dimer interface. The positively and
negatively charged areas are colored blue and red, respectively.
All ligands for both monomers are shown as dark yellow-colored
space-filling representations. The side chains that are only
conserved among bacterial NOSs are shown as balls and sticks,
with the nitrogen and oxygen atoms colored in blue and red,
respectively.(C) Stereo view of one set of ligand binding sites
of SANOS. The main chain backbone of the A and B chains are
colored dark and light gray, respectively. Haem, SEITU, and the
nicotinamide and ribose moieties of NAD^+ are colored by atoms,
with carbon atoms in dark gray. The haem iron is shown as a
magenta sphere. The side chains of key residues are drawn as
ball-and-stick representations and colored by atoms, with their
carbon atoms in cyan. Water molecules are represented as red
spheres. The broken yellow lines indicate hydrogen bonds between
the substrates and the protein. SEITU, H[4]B, and a section of
the hook from bovine eNOS that interacts with the pterin
(colored orange) have been overlaid onto the SANOS interface
ligand binding site.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2002,
10,
1687-1696)
copyright 2002.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Maréchal,
T.A.Mattioli,
D.J.Stuehr,
and
J.Santolini
(2010).
NO synthase isoforms specifically modify peroxynitrite reactivity.
|
| |
FEBS J,
277,
3963-3973.
|
 |
|
|
|
|
 |
B.R.Crane,
J.Sudhamsu,
and
B.A.Patel
(2010).
Bacterial nitric oxide synthases.
|
| |
Annu Rev Biochem,
79,
445-470.
|
 |
|
|
|
|
 |
J.Sudhamsu,
and
B.R.Crane
(2009).
Bacterial nitric oxide synthases: what are they good for?
|
| |
Trends Microbiol,
17,
212-218.
|
 |
|
|
|
|
 |
I.Gusarov,
M.Starodubtseva,
Z.Q.Wang,
L.McQuade,
S.J.Lippard,
D.J.Stuehr,
and
E.Nudler
(2008).
Bacterial nitric-oxide synthases operate without a dedicated redox partner.
|
| |
J Biol Chem,
283,
13140-13147.
|
 |
|
|
|
|
 |
F.J.Chartier,
and
M.Couture
(2007).
Substrate-specific interactions with the heme-bound oxygen molecule of nitric-oxide synthase.
|
| |
J Biol Chem,
282,
20877-20886.
|
 |
|
|
|
|
 |
Z.Q.Wang,
R.J.Lawson,
M.R.Buddha,
C.C.Wei,
B.R.Crane,
A.W.Munro,
and
D.J.Stuehr
(2007).
Bacterial flavodoxins support nitric oxide production by Bacillus subtilis nitric-oxide synthase.
|
| |
J Biol Chem,
282,
2196-2202.
|
 |
|
|
|
|
 |
R.Loria,
J.Kers,
and
M.Joshi
(2006).
Evolution of plant pathogenicity in Streptomyces.
|
| |
Annu Rev Phytopathol,
44,
469-487.
|
 |
|
|
|
|
 |
D.J.Stuehr,
J.Santolini,
Z.Q.Wang,
C.C.Wei,
and
S.Adak
(2004).
Update on mechanism and catalytic regulation in the NO synthases.
|
| |
J Biol Chem,
279,
36167-36170.
|
 |
|
|
|
|
 |
F.J.Chartier,
and
M.Couture
(2004).
Stability of the heme environment of the nitric oxide synthase from Staphylococcus aureus in the absence of pterin cofactor.
|
| |
Biophys J,
87,
1939-1950.
|
 |
|
|
|
|
 |
M.R.Buddha,
T.Tao,
R.J.Parry,
and
B.R.Crane
(2004).
Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase.
|
| |
J Biol Chem,
279,
49567-49570.
|
 |
|
|
|
|
 |
I.S.Hong,
Y.K.Kim,
W.S.Choi,
D.W.Seo,
J.W.Yoon,
J.W.Han,
H.Y.Lee,
and
H.W.Lee
(2003).
Purification and characterization of nitric oxide synthase from Staphylococcus aureus.
|
| |
FEMS Microbiol Lett,
222,
177-182.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

