 |
PDBsum entry 1gc2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of the pyridoxal-5'-phosphate dependent l-methionine gamma-lyase from pseudomonas putida
|
|
Structure:
|
 |
Methionine gamma-lyase. Chain: a, b, c, d. Ec: 4.4.1.11
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.211
|
R-free:
|
0.243
|
|
|
Authors:
|
 |
H.Motoshima,K.Inagaki,T.Kumasaka,M.Furuichi,H.Inoue,T.Tamura,N.Esaki, K.Soda,N.Tanaka,M.Yamamoto,H.Tanaka
|
|
Key ref:
|
 |
H.Motoshima
et al.
(2000).
Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J Biochem (tokyo),
128,
349-354.
PubMed id:
|
 |
|
Date:
|
 |
|
06-Jul-00
|
Release date:
|
08-May-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P13254
(MEGL_PSEPU) -
L-methionine gamma-lyase from Pseudomonas putida
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
398 a.a.
373 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.4.1.11
- methionine gamma-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionine + H2O = methanethiol + 2-oxobutanoate + NH4+
|
 |
 |
 |
 |
 |
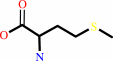 L-methionine
L-methionine
|
+
|
H2O
|
=
|
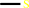 methanethiol
methanethiol
|
+
|
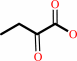 2-oxobutanoate
2-oxobutanoate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.2
- homocysteine desulfhydrase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homocysteine + H2O = 2-oxobutanoate + hydrogen sulfide + NH4+ + H+
|
 |
 |
 |
 |
 |
 L-homocysteine
L-homocysteine
|
+
|
H2O
|
=
|
 2-oxobutanoate
2-oxobutanoate
|
+
|
hydrogen sulfide
|
+
|
NH4(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biochem (tokyo)
128:349-354
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
|
|
H.Motoshima,
K.Inagaki,
T.Kumasaka,
M.Furuichi,
H.Inoue,
T.Tamura,
N.Esaki,
K.Soda,
N.Tanaka,
M.Yamamoto,
H.Tanaka.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
L-Methionine gamma-lyase (MGL) catalyzes the pyridoxal 5'-phosphate (PLP)
dependent alpha,gamma-elimination of L-methionine. We have determined two
crystal structures of MGL from Pseudomonas putida using MAD (multiwavelength
anomalous diffraction) and molecular replacement methods. The structures have
been refined to an R-factor of 21.1% at 2.0 and 1.7 A resolution using
synchrotron radiation diffraction data. A homotetramer with 222 symmetry is
built up by non-crystallographic symmetry. Two monomers associate to build the
active dimer. The spatial fold of subunits, with three functionally distinct
domains and their quarternary arrangement, is similar to those of
L-cystathionine beta-lyase and L-cystathionine gamma-synthase from Escherichia
coli.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.S.El-Sayed
(2010).
Microbial L-methioninase: production, molecular characterization, and therapeutic applications.
|
| |
Appl Microbiol Biotechnol,
86,
445-467.
|
 |
|
|
|
|
 |
A.Nikulin,
S.Revtovich,
E.Morozova,
N.Nevskaya,
S.Nikonov,
M.Garber,
and
T.Demidkina
(2008).
High-resolution structure of methionine gamma-lyase from Citrobacter freundii.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
211-218.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Sato,
T.Karaki,
A.Shimizu,
K.Kamei,
S.Harada,
and
T.Nozaki
(2008).
Crystallization and preliminary X-ray analysis of L-methionine gamma-lyase 1 from Entamoeba histolytica.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
697-699.
|
 |
|
|
|
|
 |
A.Goyer,
E.Collakova,
Y.Shachar-Hill,
and
A.D.Hanson
(2007).
Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway.
|
| |
Plant Cell Physiol,
48,
232-242.
|
 |
|
|
|
|
 |
V.Ali,
and
T.Nozaki
(2007).
Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by "amitochondriate" protozoan parasites.
|
| |
Clin Microbiol Rev,
20,
164-187.
|
 |
|
|
|
|
 |
D.Sato,
W.Yamagata,
K.Kamei,
T.Nozaki,
and
S.Harada
(2006).
Expression, purification and crystallization of L-methionine gamma-lyase 2 from Entamoeba histolytica.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1034-1036.
|
 |
|
|
|
|
 |
I.V.Manukhov,
D.V.Mamaeva,
E.A.Morozova,
S.M.Rastorguev,
N.G.Faleev,
T.V.Demidkina,
and
G.B.Zavilgelsky
(2006).
L-methionine gamma-lyase from Citrobacter freundii: cloning of the gene and kinetic parameters of the enzyme.
|
| |
Biochemistry (Mosc),
71,
361-369.
|
 |
|
|
|
|
 |
T.Takakura,
T.Ito,
S.Yagi,
Y.Notsu,
T.Itakura,
T.Nakamura,
K.Inagaki,
N.Esaki,
R.M.Hoffman,
and
A.Takimoto
(2006).
High-level expression and bulk crystallization of recombinant L-methionine gamma-lyase, an anticancer agent.
|
| |
Appl Microbiol Biotechnol,
70,
183-192.
|
 |
|
|
|
|
 |
D.V.Mamaeva,
E.A.Morozova,
A.D.Nikulin,
S.V.Revtovich,
S.V.Nikonov,
M.B.Garber,
and
T.V.Demidkina
(2005).
Structure of Citrobacter freundii L-methionine gamma-lyase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
546-549.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Amarita,
M.Yvon,
M.Nardi,
E.Chambellon,
J.Delettre,
and
P.Bonnarme
(2004).
Identification and functional analysis of the gene encoding methionine-gamma-lyase in Brevibacterium linens.
|
| |
Appl Environ Microbiol,
70,
7348-7354.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

