 |
PDBsum entry 1dao

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Flavoenzyme
|
 |
|
Title:
|
 |
Covalent adduct of d-amino acid oxidase from pig kidney with 3-methyl- 2-oxo-valeric acid
|
|
Structure:
|
 |
D-amino acid oxidase. Chain: a, b, c, d, e, f, g, h. Synonym: daao. Ec: 1.4.3.3
|
|
Source:
|
 |
Sus scrofa. Pig. Organism_taxid: 9823. Organ: kidney. Organelle: peroxisome
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
3.20Å
|
R-factor:
|
0.232
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
F.Todone,A.Mattevi
|
Key ref:

|
 |
F.Todone
et al.
(1997).
Active site plasticity in D-amino acid oxidase: a crystallographic analysis.
Biochemistry,
36,
5853-5860.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Jan-97
|
Release date:
|
23-Jul-97
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00371
(OXDA_PIG) -
D-amino-acid oxidase from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
347 a.a.
339 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.4.3.3
- D-amino-acid oxidase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Cephalosporin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a D-alpha-amino acid + O2 + H2O = a 2-oxocarboxylate + H2O2 + NH4+
|
 |
 |
 |
 |
 |
D-alpha-amino acid
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
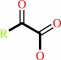 2-oxocarboxylate
2-oxocarboxylate
|
+
|
 H2O2
H2O2
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAB)
matches with 91.38% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:5853-5860
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Active site plasticity in D-amino acid oxidase: a crystallographic analysis.
|
|
F.Todone,
M.A.Vanoni,
A.Mozzarelli,
M.Bolognesi,
A.Coda,
B.Curti,
A.Mattevi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
D-Amino acid oxidase (DAAO) is the prototype of the flavin-containing oxidases.
It catalyzes the oxidative deamination of various D-amino acids, ranging from
D-Ala to D-Trp. We have carried out the X-ray analysis of reduced DAAO in
complex with the reaction product imino tryptophan (iTrp) and of the covalent
adduct generated by the photoinduced reaction of the flavin with
3-methyl-2-oxobutyric acid (kVal). These structures were solved by combination
of 8-fold density averaging and least-squares refinement techniques. The FAD
redox state of DAAO crystals was assessed by single-crystal polarized absorption
microspectrophotometry. iTrp binds to the reduced enzyme with the N, C alpha, C,
and C beta atoms positioned 3.8 A from the re side of the flavin. The indole
side chain points away from the cofactor and is bound in the active site through
a rotation of Tyr224. This residue plays a crucial role in that it adapts its
conformation to the size of the active site ligand, providing the enzyme with
the plasticity required for binding a broad range of substrates. The iTrp
binding mode is fully consistent with the proposal, inferred from the analysis
of the native DAAO structure, that substrate oxidation occurs via direct hydride
transfer from the C alpha to the flavin N5 atom. In this regard, it is
remarkable that, even in the presence of the bulky iTrp ligand, the active
center is made solvent inaccessible by loop 216-228. This loop is thought to
switch between the "closed" conformation observed in the crystal structures and
an "open" state required for substrate binding and product release. Loop closure
is likely to have a role in catalysis by increasing the hydrophobicity of the
active site, thus making the hydride transfer reaction more effective. Binding
of kVal leads to keto acid decarboxylation and formation of a covalent bond
between the keto acid C alpha and the flavin N5 atoms. Formation of this acyl
adduct results in a nonplanar flavin, characterized by a 22 degrees angle
between the pyrimidine and benzene rings. Thus, in addition to an adaptable
substrate binding site, DAAO has the ability to bind a highly distorted
cofactor. This ability is relevant for the enzyme's function as a highly
efficient oxidase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Katane,
Y.Saitoh,
K.Maeda,
T.Hanai,
M.Sekine,
T.Furuchi,
and
H.Homma
(2011).
Role of the active site residues arginine-216 and arginine-237 in the substrate specificity of mammalian D-aspartate oxidase.
|
| |
Amino Acids,
40,
467-476.
|
 |
|
|
|
|
 |
J.Mitchell,
P.Paul,
H.J.Chen,
A.Morris,
M.Payling,
M.Falchi,
J.Habgood,
S.Panoutsou,
S.Winkler,
V.Tisato,
A.Hajitou,
B.Smith,
C.Vance,
C.Shaw,
N.D.Mazarakis,
and
J.de Belleroche
(2010).
Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase.
|
| |
Proc Natl Acad Sci U S A,
107,
7556-7561.
|
 |
|
|
|
|
 |
L.Liu,
J.F.Wu,
Y.F.Ma,
S.Y.Wang,
G.P.Zhao,
and
S.J.Liu
(2007).
A novel deaminase involved in chloronitrobenzene and nitrobenzene degradation with Comamonas sp. strain CNB-1.
|
| |
J Bacteriol,
189,
2677-2682.
|
 |
|
|
|
|
 |
M.Katane,
Y.Seida,
M.Sekine,
T.Furuchi,
and
H.Homma
(2007).
Caenorhabditis elegans has two genes encoding functional d-aspartate oxidases.
|
| |
FEBS J,
274,
137-149.
|
 |
|
|
|
|
 |
T.Kawazoe,
H.Tsuge,
M.S.Pilone,
and
K.Fukui
(2006).
Crystal structure of human D-amino acid oxidase: context-dependent variability of the backbone conformation of the VAAGL hydrophobic stretch located at the si-face of the flavin ring.
|
| |
Protein Sci,
15,
2708-2717.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Abe,
N.Yoshikawa,
M.G.Sarower,
and
S.Okada
(2005).
Physiological function and metabolism of free D-alanine in aquatic animals.
|
| |
Biol Pharm Bull,
28,
1571-1577.
|
 |
|
|
|
|
 |
V.I.Tishkov,
and
S.V.Khoronenkova
(2005).
D-Amino acid oxidase: structure, catalytic mechanism, and practical application.
|
| |
Biochemistry (Mosc),
70,
40-54.
|
 |
|
|
|
|
 |
S.J.Teague
(2003).
Implications of protein flexibility for drug discovery.
|
| |
Nat Rev Drug Discov,
2,
527-541.
|
 |
|
|
|
|
 |
A.Boselli,
S.Sacchi,
V.Job,
M.S.Pilone,
and
L.Pollegioni
(2002).
Role of tyrosine 238 in the active site of Rhodotorula gracilis D-amino acid oxidase. A site-directed mutagenesis study.
|
| |
Eur J Biochem,
269,
4762-4771.
|
 |
|
|
|
|
 |
C.Binda,
R.Angelini,
R.Federico,
P.Ascenzi,
and
A.Mattevi
(2001).
Structural bases for inhibitor binding and catalysis in polyamine oxidase.
|
| |
Biochemistry,
40,
2766-2776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Breithaupt,
J.Strassner,
U.Breitinger,
R.Huber,
P.Macheroux,
A.Schaller,
and
T.Clausen
(2001).
X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
|
| |
Structure,
9,
419-429.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.S.Park,
and
H.S.Kim
(2001).
Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process.
|
| |
J Bacteriol,
183,
5074-5081.
|
 |
|
|
|
|
 |
R.Miura
(2001).
Versatility and specificity in flavoenzymes: control mechanisms of flavin reactivity.
|
| |
Chem Rec,
1,
183-194.
|
 |
|
|
|
|
 |
Y.Liu,
T.M.Louie,
J.Payne,
J.Bohuslavek,
H.Bolton,
and
L.Xun
(2001).
Identification, purification, and characterization of iminodiacetate oxidase from the EDTA-degrading bacterium BNC1.
|
| |
Appl Environ Microbiol,
67,
696-701.
|
 |
|
|
|
|
 |
P.D.Pawelek,
J.Cheah,
R.Coulombe,
P.Macheroux,
S.Ghisla,
and
A.Vrielink
(2000).
The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
|
| |
EMBO J,
19,
4204-4215.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Umhau,
L.Pollegioni,
G.Molla,
K.Diederichs,
W.Welte,
M.S.Pilone,
and
S.Ghisla
(2000).
The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
|
| |
Proc Natl Acad Sci U S A,
97,
12463-12468.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.M.Harris,
G.Molla,
M.S.Pilone,
and
L.Pollegioni
(1999).
Studies on the reaction mechanism of Rhodotorula gracilis D-amino-acid oxidase. Role of the highly conserved Tyr-223 on substrate binding and catalysis.
|
| |
J Biol Chem,
274,
36233-36240.
|
 |
|
|
|
|
 |
A.Mattevi
(1998).
The PHBH fold: not only flavoenzymes.
|
| |
Biophys Chem,
70,
217-222.
|
 |
|
|
|
|
 |
D.Parsonage,
J.Luba,
T.C.Mallett,
and
A.Claiborne
(1998).
The soluble alpha-glycerophosphate oxidase from Enterococcus casseliflavus. Sequence homology with the membrane-associated dehydrogenase and kinetic analysis of the recombinant enzyme.
|
| |
J Biol Chem,
273,
23812-23822.
|
 |
|
|
|
|
 |
Z.He,
and
J.C.Spain
(1998).
A novel 2-aminomuconate deaminase in the nitrobenzene degradation pathway of Pseudomonas pseudoalcaligenes JS45.
|
| |
J Bacteriol,
180,
2502-2506.
|
 |
|
|
|
|
 |
A.Mattevi,
M.A.Vanoni,
and
B.Curti
(1997).
Structure of D-amino acid oxidase: new insights from an old enzyme.
|
| |
Curr Opin Struct Biol,
7,
804-810.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

