Assemblies
Assembly Name:
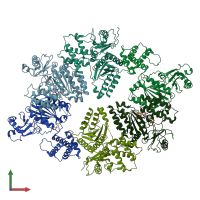
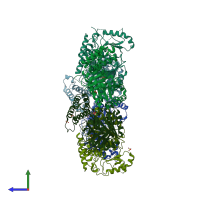
Putative pachytene checkpoint protein 2
Multimeric state:
homo hexamer
Accessible surface area:
102721.83 Å2
Buried surface area:
17324.62 Å2
Dissociation area:
4,435.82
Å2
Dissociation energy (ΔGdiss):
5.66
kcal/mol
Dissociation entropy (TΔSdiss):
45.83
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-170812
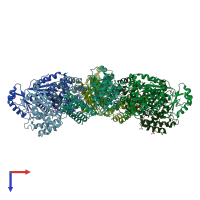
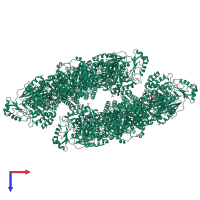
Assembly Name:
Putative pachytene checkpoint protein 2
Multimeric state:
homo dodecamer
Accessible surface area:
203858.74 Å2
Buried surface area:
36234.16 Å2
Dissociation area:
9,664.09
Å2
Dissociation energy (ΔGdiss):
1.33
kcal/mol
Dissociation entropy (TΔSdiss):
109.42
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-170811
Macromolecules
Chains: A, B, C, D, E, F
Length: 424 amino acids
Theoretical weight: 48.08 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
Pfam: ATPase family associated with various cellular activities (AAA)
InterPro:
Length: 424 amino acids
Theoretical weight: 48.08 KDa
Source organism: Caenorhabditis elegans
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q09535 (Residues: 1-424; Coverage: 100%)
Q09535 (Residues: 1-424; Coverage: 100%)
Pfam: ATPase family associated with various cellular activities (AAA)
InterPro: