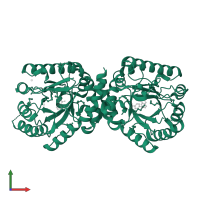
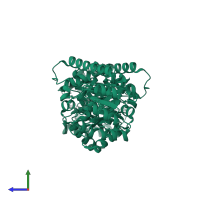
Assemblies
Assembly Name:
dihydropteroate synthase
Multimeric state:
homo tetramer
Accessible surface area:
45350.11 Å2
Buried surface area:
6072.55 Å2
Dissociation area:
1,294.94
Å2
Dissociation energy (ΔGdiss):
-26.92
kcal/mol
Dissociation entropy (TΔSdiss):
39.74
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-182914
Assembly Name:
dihydropteroate synthase
Multimeric state:
homo dimer
Accessible surface area:
21090.47 Å2
Buried surface area:
4738.76 Å2
Dissociation area:
1,413.11
Å2
Dissociation energy (ΔGdiss):
17.1
kcal/mol
Dissociation entropy (TΔSdiss):
13.25
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-182913
Assembly Name:
dihydropteroate synthase
Multimeric state:
homo dimer
Accessible surface area:
21159.14 Å2
Buried surface area:
4442.24 Å2
Dissociation area:
1,437.3
Å2
Dissociation energy (ΔGdiss):
16.15
kcal/mol
Dissociation entropy (TΔSdiss):
13.26
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-182913
Macromolecules
Chains: A, B, C, D
Length: 314 amino acids
Theoretical weight: 35.88 KDa
Source organism: Coxiella burnetii
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Pterin binding enzyme
InterPro:
Length: 314 amino acids
Theoretical weight: 35.88 KDa
Source organism: Coxiella burnetii
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q83BY6 (Residues: 1-297; Coverage: 100%)
Q83BY6 (Residues: 1-297; Coverage: 100%)
Pfam: Pterin binding enzyme
InterPro:
- Dihydropteroate synthase-like superfamily
- Dihydropteroate synthase-like
- Pterin-binding domain
- Dihydropteroate synthase domain