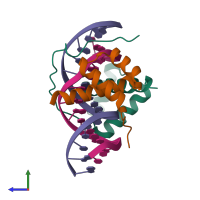
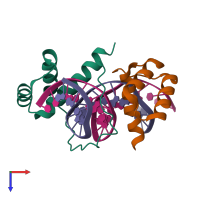
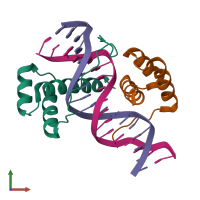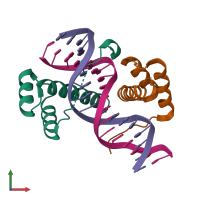Assemblies
Multimeric state:
hetero tetramer
Accessible surface area:
13027.62 Å2
Buried surface area:
6775.37 Å2
Dissociation area:
1,303.78
Å2
Dissociation energy (ΔGdiss):
8.4
kcal/mol
Dissociation entropy (TΔSdiss):
10.9
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-116053
Multimeric state:
hetero tetramer
Accessible surface area:
12615.06 Å2
Buried surface area:
6188.55 Å2
Dissociation area:
1,160.86
Å2
Dissociation energy (ΔGdiss):
8.73
kcal/mol
Dissociation entropy (TΔSdiss):
10.99
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-116053
Macromolecules
Chains: A, E
Length: 93 amino acids
Theoretical weight: 11.19 KDa
Source organism: Drosophila melanogaster
Expression system: Escherichia coli
UniProt:
Pfam: Homeodomain
InterPro:
Length: 93 amino acids
Theoretical weight: 11.19 KDa
Source organism: Drosophila melanogaster
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q9VD99 (Residues: 169-261; Coverage: 30%)
Q9VD99 (Residues: 169-261; Coverage: 30%)
Pfam: Homeodomain
InterPro:
- T-cell leukemia homeobox protein 1/2/3
- Homeobox-like domain superfamily
- Homeobox domain
- Homeobox domain, metazoa
- Homeobox, conserved site
Chains: B, F
Length: 67 amino acids
Theoretical weight: 8.34 KDa
Source organism: Drosophila melanogaster
Expression system: Escherichia coli
UniProt:
Pfam: Homeodomain
InterPro:
Length: 67 amino acids
Theoretical weight: 8.34 KDa
Source organism: Drosophila melanogaster
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q06453 (Residues: 80-146; Coverage: 16%)
Q06453 (Residues: 80-146; Coverage: 16%)
Pfam: Homeodomain
InterPro:
- Homeobox-like domain superfamily
- Homeobox domain
- Homeobox domain, metazoa
- Homeobox, conserved site
- Helix-turn-helix motif
Name:
5'-D(*GP*GP*CP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*GP*CP*GP*G)-3'
Representative chains: C, G
Length: 17 nucleotides
Theoretical weight: 5.26 KDa
Representative chains: C, G
Length: 17 nucleotides
Theoretical weight: 5.26 KDa
Name:
5'-D(*CP*CP*GP*CP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*GP*CP*C)-3'
Representative chains: D, H
Length: 17 nucleotides
Theoretical weight: 5.16 KDa
Representative chains: D, H
Length: 17 nucleotides
Theoretical weight: 5.16 KDa