Adenylosuccinate lyase
Adenylosuccinate lyase catalyses two similar but separate reactions in the de novo purine synthesis pathway. In the first reaction is converts 5-aminoimidazole-(N-succinylcarboxyamide) ribotide into 5-aminoimidazole-4-carboxyamide ribotide in the ninth step of the synthesis of inosine monophosphate. In the second reaction the enzyme converts adenylosuccinate into adenosine monophospate which occurs four steps after the first reaction. Adenylosuccinate lyase helps provide the majority of purine nucleotides required for DNA replication as well as playing a role in cellular metabolism as an enzyme in the purine nucleotide cycle. The purine nucleotide cycle controls both the amounts of available citric acid intermediates and the amount of free AMP. Mutations in the enzyme leads to severe clinical consequences including mental retardation with autistic features.
Reference Protein and Structure
- Sequence
-
Q9X0I0
 (4.3.2.2)
(4.3.2.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermotoga maritima MSB8 (Bacteria)

- PDB
-
1c3c
- T. MARITIMA ADENYLOSUCCINATE LYASE
(1.8 Å)



- Catalytic CATH Domains
-
1.10.275.10
 1.20.200.10
1.20.200.10  (see all for 1c3c)
(see all for 1c3c)
- Cofactors
- Water (1)
Enzyme Reaction (EC:4.3.2.2)
Enzyme Mechanism
Introduction
The catalytic reaction proceeds via a general acid/base mechanism in which the C-beta proton of the substrate is abstracted by the general base (thought to be serine), yielding a carbanion intermediate. This step is followed by proton donation by a catalytic acid (thought to be histidine), resulting in C-N bond cleavage and product release. Recent studies on Plasmodium falciparum have suggested that the C-N bond cleavage is the rate-limiting step. Thought to proceed via a uni-bi mechanism kinetic mechanism.
Catalytic Residues Roles
| UniProt | PDB* (1c3c) | ||
| Ser263 | Ser263(262)B | Acts as the general acid/base during the course of the reaction. It is not yet clear how this residue is stabilised/activated to act as the general base in the first proton abstraction reaction, partly due to its absence in the crystal structure. Should actually be Ser262, but this residue is missing in the PDB file. | proton acceptor, proton donor |
| Glu275 | Glu275(274)B | The role of this glutamate is to prime the general acid/base histidine for function as a catalytic acid; this charge-relay interaction is conserved in other superfamily members. | increase basicity, activator, hydrogen bond acceptor |
| His141 | His141(140)A | The position of the side chain of this histidine reveals that it functions as a catalytic acid via a charge-relay interaction with a glutamate residue that is similarly conserved across the ASL superfamily. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Lys268, Thr140 | Lys268(267)B, Thr140(139)A | Aid in stabilising the reactive intermediates and transition states. | electrostatic stabiliser |
| His68 | His68(67)A(AA) | No longer thought to be the general acid/base. Thougt to be involved in binding and and stabilisng one of the carboxylate groups of the substrate's succinyl moiety. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular elimination, rate-determining step, inferred reaction stepReferences
- Toth EA et al. (2000), Structure, 8, 163-174. The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway. DOI:10.1016/s0969-2126(00)00092-7. PMID:10673438.
- Puthan Veetil V et al. (2012), Biochemistry, 51, 4237-4243. Aspartase/Fumarase Superfamily: A Common Catalytic Strategy Involving General Base-Catalyzed Formation of a Highly Stabilizedaci-Carboxylate Intermediate. DOI:10.1021/bi300430j. PMID:22551392.
- Fyfe PK et al. (2010), Acta Crystallogr D Biol Crystallogr, 66, 881-888. Structure ofStaphylococcus aureusadenylosuccinate lyase (PurB) and assessment of its potential as a target for structure-based inhibitor discovery. DOI:10.1107/s0907444910020081. PMID:20693687.
- Kozlov G et al. (2009), Acta Crystallogr Sect F Struct Biol Cryst Commun, 65, 857-861. The structure of phosphate-boundEscherichia coliadenylosuccinate lyase identifies His171 as a catalytic acid. DOI:10.1107/s1744309109029674. PMID:19724117.
- Bulusu V et al. (2009), Biochim Biophys Acta, 1794, 642-654. Elucidation of the substrate specificity, kinetic and catalytic mechanism of adenylosuccinate lyase from Plasmodium falciparum. DOI:10.1016/j.bbapap.2008.11.021. PMID:19111634.
- Sivendran S et al. (2008), Protein Sci, 17, 1162-1174. Effect of a new non-cleavable substrate analog on wild-type and serine mutants in the signature sequence of adenylosuccinate lyase ofBacillus subtilisandHomo sapiens. DOI:10.1110/ps.034777.108. PMID:18469177.
- Tsai M et al. (2007), J Mol Biol, 370, 541-554. Substrate and Product Complexes of Escherichia coli Adenylosuccinate Lyase Provide New Insights into the Enzymatic Mechanism. DOI:10.1016/j.jmb.2007.04.052. PMID:17531264.
- Segall ML et al. (2007), Protein Sci, 16, 441-448. Important roles of hydroxylic amino acid residues in the function of Bacillus subtilis adenylosuccinate lyase. DOI:10.1110/ps.062650007. PMID:17322529.
- Sivendran S et al. (2007), Protein Sci, 16, 1700-1707. Effect of Asp69and Arg310on the pK of His68, a key catalytic residue of adenylosuccinate lyase. DOI:10.1110/ps.072927207. PMID:17600142.
- Bhaumik P et al. (2004), Acta Crystallogr D Biol Crystallogr, 60, 1964-1970. Structure determination and refinement at 2.44 Å resolution of argininosuccinate lyase fromEscherichia coli. DOI:10.1107/s0907444904021912. PMID:15502303.
- Segall ML et al. (2004), Biochemistry, 43, 7391-7402. Gln212, Asn270, and Arg301Are Critical for Catalysis by Adenylosuccinate Lyase fromBacillus subtilis†. DOI:10.1021/bi0494774. PMID:15182182.
- Brosius JL et al. (2002), Biochemistry, 41, 2217-2226. Three subunits contribute amino acids to the active site of tetrameric adenylosuccinate lyase: Lys268 and Glu275 are required. PMID:11841213.
- Weaver TM et al. (1995), Nat Struct Biol, 2, 654-662. The multisubunit active site of fumarase C from Escherichia coli. PMID:7552727.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu275(274)B | hydrogen bond acceptor |
| His141(140)A | hydrogen bond acceptor, hydrogen bond donor |
| Ser263(262)B | proton donor |
Chemical Components
proton transfer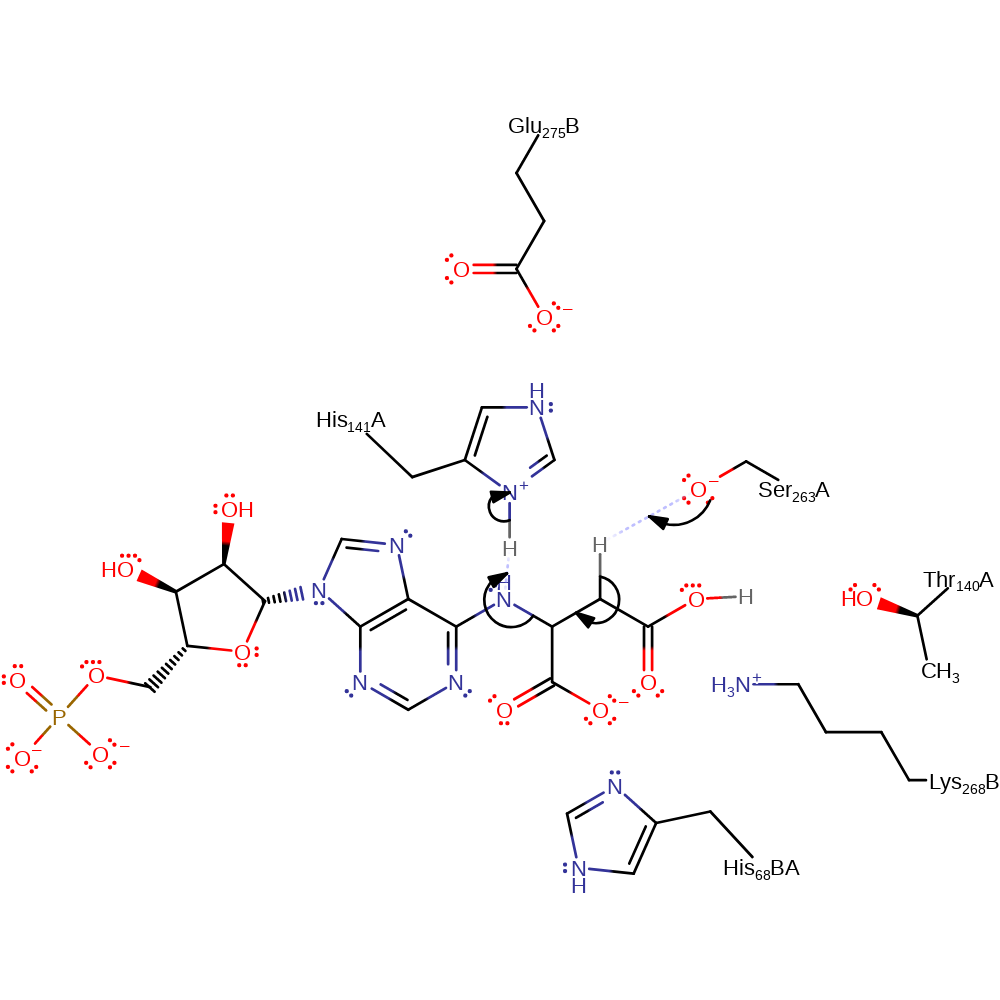
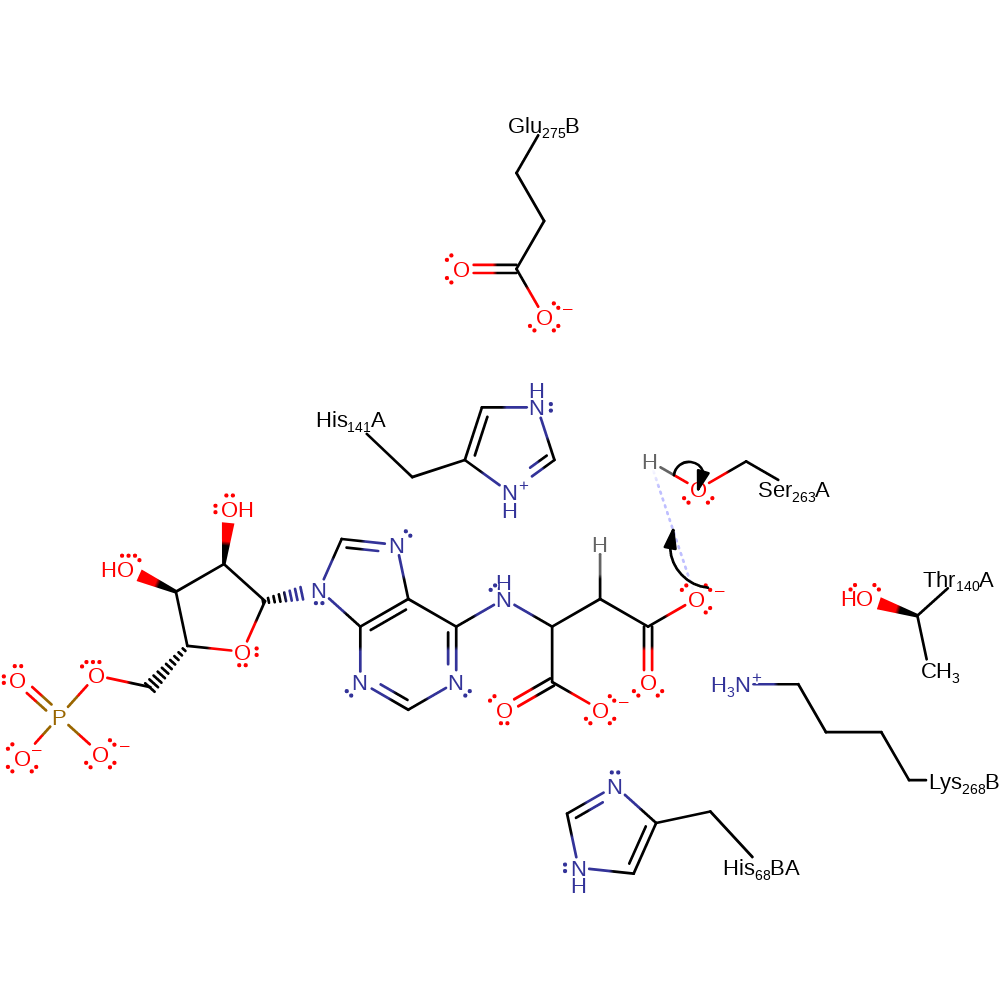
Step 2. Ser263 deprotonates the substrate, initiating the elimination of AMP, which deprotonates His141. This is thought to be the rate determining step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His141(140)A | hydrogen bond donor |
| Glu275(274)B | activator |
| Thr140(139)A | electrostatic stabiliser |
| Lys268(267)B | electrostatic stabiliser |
| His68(67)A(AA) | electrostatic stabiliser |
| Ser263(262)B | proton acceptor |
| His141(140)A | proton donor |
Chemical Components
ingold: bimolecular elimination, proton transfer, rate-determining stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu275(274)B | increase basicity |
| His141(140)A | proton acceptor |



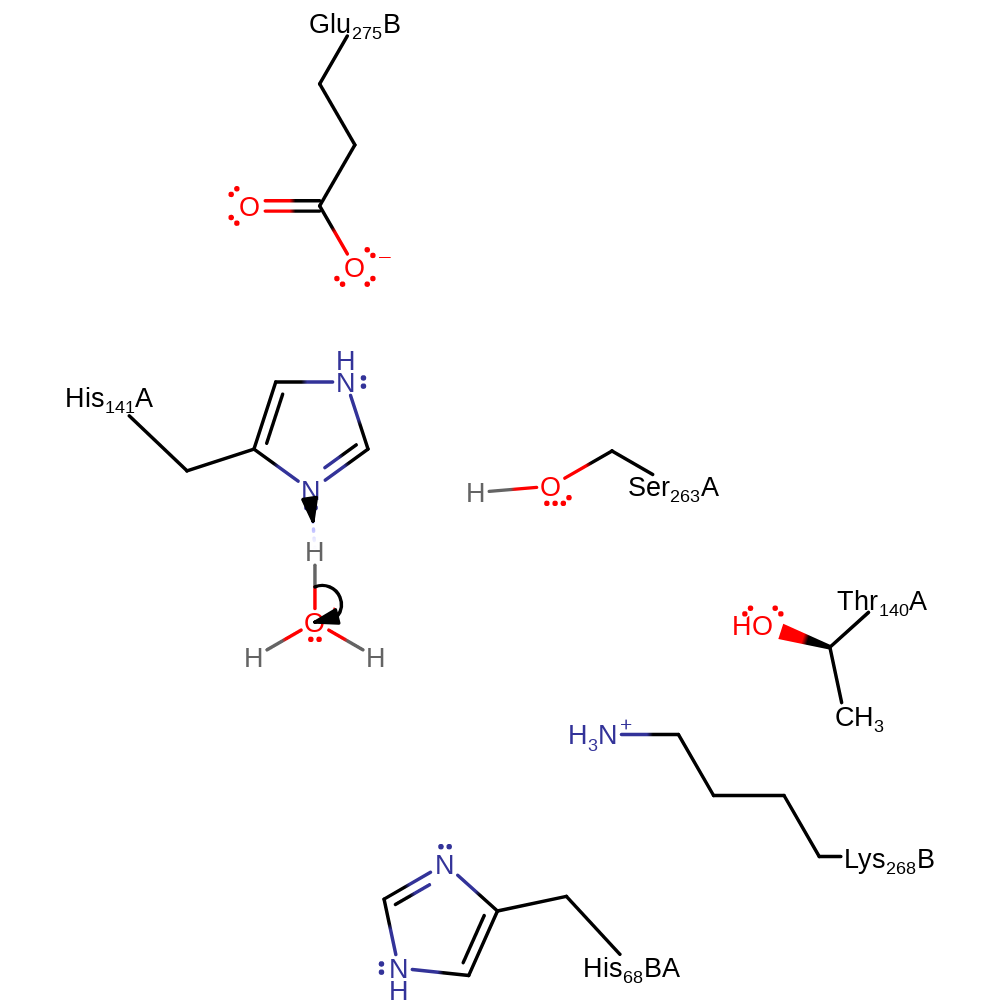
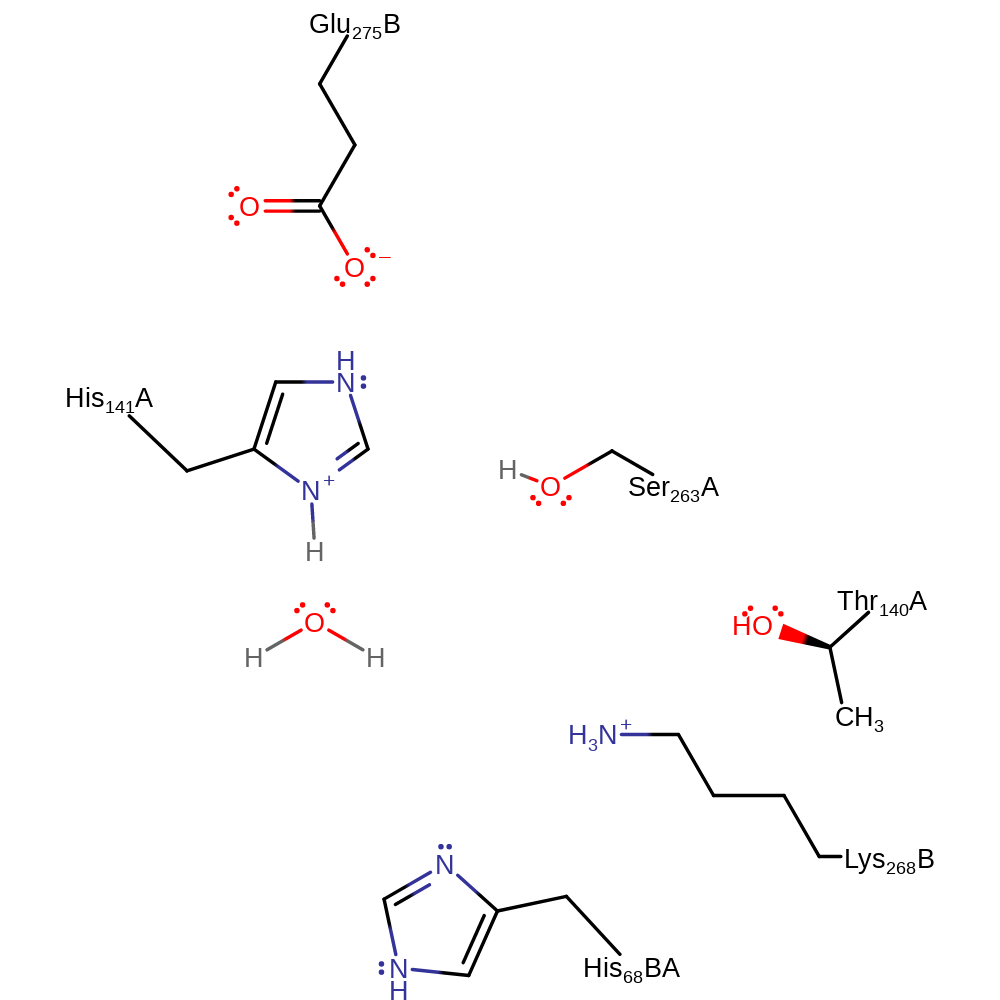 Download:
Download: