 |
PDBsum entry 1c3c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.2.2
- adenylosuccinate lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N6-(1,2-dicarboxyethyl)-AMP = fumarate + AMP
|
|
2.
|
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate = 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamide + fumarate
|
|
 |
 |
 |
 |
 |
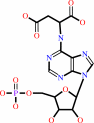 N(6)-(1,2-dicarboxyethyl)-AMP
N(6)-(1,2-dicarboxyethyl)-AMP
|
=
|
 fumarate
fumarate
|
+
|
 AMP
AMP
|
|
 |
 |
 |
 |
 |
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate
|
=
|
5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamide
|
+
|
 fumarate
fumarate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
8:163-174
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
|
|
E.A.Toth,
T.O.Yeates.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Background: Adenylosuccinate lyase is an enzyme that plays a critical role in
both cellular replication and metabolism via its action in the de novo purine
biosynthetic pathway. Adenylosuccinate lyase is the only enzyme in this pathway
to catalyze two separate reactions, enabling it to participate in the addition
of a nitrogen at two different positions in adenosine monophosphate. Both
reactions catalyzed by adenylosuccinate lyase involve the beta-elimination of
fumarate. Enzymes that catalyze this type of reaction belong to a superfamily,
the members of which are homotetramers. Because adenylosuccinate lyase plays an
integral part in maintaining proper cellular metabolism, mutations in the human
enzyme can have severe clinical consequences, including mental retardation with
autistic features. Results: The 1.8 A crystal structure of adenylosuccinate
lyase from Thermotoga maritima has been determined by multiwavelength anomalous
dispersion using the selenomethionine-substituted enzyme. The fold of the
monomer is reminiscent of other members of the beta-elimination superfamily.
However, its active tetrameric form exhibits striking differences in active-site
architecture and cleft size. Conclusions: This first structure of an
adenylosuccinate lyase reveals that, along with the catalytic base (His141) and
the catalytic acid (His68), Gln212 and Asn270 might play a vital role in
catalysis by properly orienting the succinyl moiety of the substrates. We
propose a model for the dual activity of adenylosuccinate lyase: a single 180
degrees bond rotation must occur in the substrate between the first and second
enzymatic reactions. Modeling of the pathogenic human S413P mutation indicates
that the mutation destabilizes the enzyme by disrupting the C-terminal extension.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 8.
Figure 8. Mapping of the S413P human ASL mutation to the T.
maritima ASL structure. The residue singled out by directional
profiles (Asp406) is colored green. The residue singled out by
the extensible threading calculator (ETC) method (Thr404) is
colored cyan. The C-terminal extension is colored red. This
figure was generated using RIBBONS [36].
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2000,
8,
163-174)
copyright 2000.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Yuan,
J.R.Gu,
W.B.Gu,
J.Wu,
S.R.Ge,
and
H.Xu
(2011).
Molecular cloning, characterization and expression analysis of adenylosuccinate lyase gene in grass carp (Ctenopharyngodon idella).
|
| |
Mol Biol Rep,
38,
2059-2065.
|
 |
|
|
|
|
 |
G.Allegri,
M.J.Fernandes,
F.B.Scalco,
P.Correia,
R.E.Simoni,
J.C.Llerena,
and
M.L.de Oliveira
(2010).
Fumaric aciduria: an overview and the first Brazilian case report.
|
| |
J Inherit Metab Dis,
33,
411-419.
|
 |
|
|
|
|
 |
P.K.Fyfe,
A.Dawson,
M.T.Hutchison,
S.Cameron,
and
W.N.Hunter
(2010).
Structure of Staphylococcus aureus adenylosuccinate lyase (PurB) and assessment of its potential as a target for structure-based inhibitor discovery.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
881-888.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Kozlov,
L.Nguyen,
J.Pearsall,
and
K.Gehring
(2009).
The structure of phosphate-bound Escherichia coli adenylosuccinate lyase identifies His171 as a catalytic acid.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
857-861.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Sivendran,
and
R.F.Colman
(2008).
Effect of a new non-cleavable substrate analog on wild-type and serine mutants in the signature sequence of adenylosuccinate lyase of Bacillus subtilis and Homo sapiens.
|
| |
Protein Sci,
17,
1162-1174.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
M.L.Segall,
M.A.Cashman,
and
R.F.Colman
(2007).
Important roles of hydroxylic amino acid residues in the function of Bacillus subtilis adenylosuccinate lyase.
|
| |
Protein Sci,
16,
441-448.
|
 |
|
|
|
|
 |
S.Sivendran,
M.L.Segall,
P.C.Rancy,
and
R.F.Colman
(2007).
Effect of Asp69 and Arg310 on the pK of His68, a key catalytic residue of adenylosuccinate lyase.
|
| |
Protein Sci,
16,
1700-1707.
|
 |
|
|
|
|
 |
C.Crifò,
W.Siems,
S.Soro,
and
C.Salerno
(2005).
Inhibition of defective adenylosuccinate lyase by HNE: a neurological disease that may be affected by oxidative stress.
|
| |
Biofactors,
24,
131-136.
|
 |
|
|
|
|
 |
P.Bhaumik,
M.K.Koski,
U.Bergmann,
and
R.K.Wierenga
(2004).
Structure determination and refinement at 2.44 A resolution of argininosuccinate lyase from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1964-1970.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Sivendran,
D.Patterson,
E.Spiegel,
I.McGown,
D.Cowley,
and
R.F.Colman
(2004).
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the equivalent mutant Bacillus subtilis ASL.
|
| |
J Biol Chem,
279,
53789-53797.
|
 |
|
|
|
|
 |
J.B.Palenchar,
J.M.Crocco,
and
R.F.Colman
(2003).
The characterization of mutant Bacillus subtilis adenylosuccinate lyases corresponding to severe human adenylosuccinate lyase deficiencies.
|
| |
Protein Sci,
12,
1694-1705.
|
 |
|
|
|
|
 |
J.L.Brosius,
and
R.F.Colman
(2002).
Three subunits contribute amino acids to the active site of tetrameric adenylosuccinate lyase: Lys268 and Glu275 are required.
|
| |
Biochemistry,
41,
2217-2226.
|
 |
|
|
|
|
 |
L.M.Sampaleanu,
B.Yu,
and
P.L.Howell
(2002).
Mutational analysis of duck delta 2 crystallin and the structure of an inactive mutant with bound substrate provide insight into the enzymatic mechanism of argininosuccinate lyase.
|
| |
J Biol Chem,
277,
4166-4175.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.J.Kappock,
S.E.Ealick,
and
J.Stubbe
(2000).
Modular evolution of the purine biosynthetic pathway.
|
| |
Curr Opin Chem Biol,
4,
567-572.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

