 |
PDBsum entry 6fiv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
6fiv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/hydrolase inhibitor
|
 |
|
Title:
|
 |
Structural studies of HIV and fiv proteases complexed with an efficient inhibitor of fiv pr
|
|
Structure:
|
 |
Retropepsin. Chain: a. Engineered: yes. Other_details: complexed with tl-3-093
|
|
Source:
|
 |
Feline immunodeficiency virus. Organism_taxid: 11674. Strain: (isolate petaluma). Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.155
|
R-free:
|
0.266
|
|
|
Authors:
|
 |
M.Li,T.Lee,G.Morris,G.Laco,C.Wong,A.Olson,J.Elder,A.Wlodawer, A.Gustchina
|
Key ref:

|
 |
M.Li
et al.
(2000).
Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease.
Proteins,
38,
29-40.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Dec-98
|
Release date:
|
09-Dec-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P16088
(POL_FIVPE) -
Pol polyprotein from Feline immunodeficiency virus (isolate Petaluma)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1124 a.a.
112 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.7.7.49
- RNA-directed Dna polymerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
DNA(n) + a 2'-deoxyribonucleoside 5'-triphosphate = DNA(n+1) + diphosphate
|
 |
 |
 |
 |
 |
 DNA(n)
DNA(n)
|
+
|
2'-deoxyribonucleoside 5'-triphosphate
|
=
|
DNA(n+1)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.3.1.13.2
- exoribonuclease H.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Exonucleolytic cleavage to 5'-phosphomonoester oligonucleotides in both 5'- to 3'- and 3'- to 5'-directions.
|
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.3.1.26.13
- retroviral ribonuclease H.
|
|
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
E.C.3.4.23.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
E.C.3.6.1.23
- dUTP diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dUTP + H2O = dUMP + diphosphate + H+
|
 |
 |
 |
 |
 |
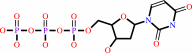 dUTP
dUTP
|
+
|
H2O
|
=
|
 dUMP
dUMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
38:29-40
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease.
|
|
M.Li,
G.M.Morris,
T.Lee,
G.S.Laco,
C.H.Wong,
A.J.Olson,
J.H.Elder,
A.Wlodawer,
A.Gustchina.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Three forms of feline immunodeficiency virus protease (FIV PR), the wild type
(wt) and two single point mutants, V59I and Q99V, as well as human
immunodeficiency virus type 1 protease (HIV-1 PR), were cocrystallized with the
C2-symmetric inhibitor, TL-3. The mutants of FIV PR were designed to replace
residues involved in enzyme-ligand interactions by the corresponding HIV-1 PR
residues at the structurally equivalent position. TL-3 shows decreased
(improved) inhibition constants with these FIV PR mutants relative to wt FIV PR.
Despite similar modes of binding of the inhibitor to all PRs (from P3 to P3'),
small differences are evident in the conformation of the Phe side chains of TL-3
at the P1 and P1' positions in the complexes with the mutated FIV PRs. The
differences mimick the observed binding of TL-3 in HIV-1 PR and correlate with a
significant improvement in the inhibition constants of TL-3 with the two mutant
FIV PRs. Large differences between the HIV-1 and FIV PR complexes are evident in
the binding modes of the carboxybenzyl groups of TL-3 at P4 and P4'. In HIV-1
PR:TL-3, these groups bind over the flap region, whereas in the FIV PR
complexes, the rings are located along the major axis of the active site. A
significant difference in the location of the flaps in this region of the HIV-1
and FIV PRs correlates with the observed conformational changes in the binding
mode of the peptidomimetic inhibitor at the P4 and P4' positions. These findings
provide a structural explanation of the observed Ki values for TL-3 with the
different PRs and will further assist in the development of improved inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. Superposition of the crystal structures of wt FIV PR
(blue) and its two mutants, V59I (yellow) and Q99V (purple),
complexed with TL-3, near the S1 binding site. Water molecules
are shown as spheres, whereas hydrogen bonds are shown as dotted
lines.
|
 |
Figure 6.
Figure 6. Comparison of the interface between the flap region
and loop 93-98 (76-81) in wt FIV PR (blue) and HIV-1 PR (pink).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2000,
38,
29-40)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Heaslet,
Y.C.Lin,
K.Tam,
B.E.Torbett,
J.H.Elder,
and
C.D.Stout
(2007).
Crystal structure of an FIV/HIV chimeric protease complexed with the broad-based inhibitor, TL-3.
|
| |
Retrovirology,
4,
1.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.C.Lin,
A.Brik,
A.de Parseval,
K.Tam,
B.E.Torbett,
C.H.Wong,
and
J.H.Elder
(2006).
Altered gag polyprotein cleavage specificity of feline immunodeficiency virus/human immunodeficiency virus mutant proteases as demonstrated in a cell-based expression system.
|
| |
J Virol,
80,
7832-7843.
|
 |
|
|
|
|
 |
A.Brik,
J.Alexandratos,
Y.C.Lin,
J.H.Elder,
A.J.Olson,
A.Wlodawer,
D.S.Goodsell,
and
C.H.Wong
(2005).
1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors.
|
| |
Chembiochem,
6,
1167-1169.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Sanches,
N.H.Martins,
A.Calazans,
R.d.e. .M.Brindeiro,
A.Tanuri,
O.A.Antunes,
and
I.Polikarpov
(2004).
Crystallization of a non-B and a B mutant HIV protease.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1625-1627.
|
 |
|
|
|
|
 |
A.Nayeem,
S.Krystek,
and
T.Stouch
(2003).
An assessment of protein-ligand binding site polarizability.
|
| |
Biopolymers,
70,
201-211.
|
 |
|
|
|
|
 |
Y.C.Lin,
Z.Beck,
G.M.Morris,
A.J.Olson,
and
J.H.Elder
(2003).
Structural basis for distinctions between substrate and inhibitor specificities for feline immunodeficiency virus and human immunodeficiency virus proteases.
|
| |
J Virol,
77,
6589-6600.
|
 |
|
|
|
|
 |
B.Bühler,
Y.C.Lin,
G.Morris,
A.J.Olson,
C.H.Wong,
D.D.Richman,
J.H.Elder,
and
B.E.Torbett
(2001).
Viral evolution in response to the broad-based retroviral protease inhibitor TL-3.
|
| |
J Virol,
75,
9502-9508.
|
 |
|
|
|
|
 |
C.C.Mak,
V.D.Le,
Y.C.Lin,
J.H.Elder,
and
C.H.Wong
(2001).
Design, synthesis, and biological evaluation of HIV/FIV protease inhibitors incorporating a conformationally constrained macrocycle with a small P3' residue.
|
| |
Bioorg Med Chem Lett,
11,
219-222.
|
 |
|
|
|
|
 |
Z.Q.Beck,
Y.C.Lin,
and
J.H.Elder
(2001).
Molecular basis for the relative substrate specificity of human immunodeficiency virus type 1 and feline immunodeficiency virus proteases.
|
| |
J Virol,
75,
9458-9469.
|
 |
|
|
|
|
 |
Y.C.Lin,
Z.Beck,
T.Lee,
V.D.Le,
G.M.Morris,
A.J.Olson,
C.H.Wong,
and
J.H.Elder
(2000).
Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease.
|
| |
J Virol,
74,
4710-4720.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

