 |
PDBsum entry 5kir

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5kir
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The structure of vioxx bound to human cox-2
|
|
Structure:
|
 |
Prostaglandin g/h synthase 2. Chain: a, b. Synonym: cyclooxygenase-2,cox-2,phs ii,prostaglandin h2 synthase 2, pghs-2,prostaglandin-endoperoxide synthase 2. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: ptgs2, cox2. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.180
|
R-free:
|
0.220
|
|
|
Authors:
|
 |
B.J.Orlando,M.G.Malkowski
|
|
Key ref:
|
 |
B.J.Orlando
and
M.G.Malkowski
(2016).
Crystal structure of rofecoxib bound to human cyclooxygenase-2.
Acta Crystallogr F Struct Biol Commun,
72,
772-776.
PubMed id:
|
 |
|
Date:
|
 |
|
16-Jun-16
|
Release date:
|
28-Sep-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P35354
(PGH2_HUMAN) -
Prostaglandin G/H synthase 2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
604 a.a.
551 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.99.1
- prostaglandin-endoperoxide synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(5Z,8Z,11Z,14Z)-eicosatetraenoate + AH2 + 2 O2 = prostaglandin H2 + A + H2O
|
 |
 |
 |
 |
 |
(5Z,8Z,11Z,14Z)-eicosatetraenoate
|
+
|
AH2
|
+
|
 2
×
O2
2
×
O2
|
=
|
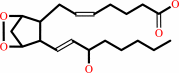 prostaglandin H2
prostaglandin H2
|
+
|
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 51.11% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr F Struct Biol Commun
72:772-776
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of rofecoxib bound to human cyclooxygenase-2.
|
|
B.J.Orlando,
M.G.Malkowski.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Rofecoxib (Vioxx) was one of the first selective cyclooxygenase-2 (COX-2)
inhibitors (coxibs) to be approved for use in humans. Within five years after
its release to the public, Vioxx was withdrawn from the market owing to the
adverse cardiovascular effects of the drug. Despite the widespread knowledge of
the development and withdrawal of Vioxx, relatively little is known at the
molecular level about how the inhibitor binds to COX-2. Vioxx is unique in that
the inhibitor contains a methyl sulfone moiety in place of the sulfonamide
moiety found in other coxibs such as celecoxib and valdecoxib. Here, new
crystallization conditions were identified that allowed the structural
determination of human COX-2 in complex with Vioxx and the structure was
subsequently determined to 2.7 Å resolution. The crystal structure provides
the first atomic level details of the binding of Vioxx to COX-2. As anticipated,
Vioxx binds with its methyl sulfone moiety located in the side pocket of the
cyclooxygenase channel, providing support for the isoform selectivity of this
drug.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

