 |
PDBsum entry 2v30

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Human orotidine 5'-phosphate decarboxylase domain of uridine monophospate synthetase (umps) in complex with its product ump.
|
|
Structure:
|
 |
Orotidine 5'-phosphate decarboxylase. Chain: a, b. Fragment: orotidine 5'-phosphate decarboxylase domain, residues 224- 479. Synonym: ompdecase, uridine monophosphate synthetase-umps. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 469008. Other_details: mammalian gene collection (mgc)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.155
|
R-free:
|
0.188
|
|
|
Authors:
|
 |
M.Moche,D.Ogg,C.Arrowsmith,H.Berglund,R.Busam,R.Collins,L.G.Dahlgren, A.Edwards,S.Flodin,A.Flores,S.Graslund,M.Hammarstrom,B.M.Hallberg, L.Holmberg-Schiavone,I.Johansson,A.Kallas,T.Karlberg,T.Kotenyova, L.Lehtio,T.Nyman,C.Persson,J.Sagemark,P.Stenmark,M.Sundstrom, A.G.Thorsell,S.Van Den Berg,J.Weigelt,M.Welin,P.Nordlund,Structural Genomics Consortium (Sgc)
|
|
Key ref:
|
 |
M.Moche
et al.
The crystal structure of human orotidine 5'- Decarboxylase domain of human uridine monophosphate synthetase (umps).
To be published,
.
|
 |
|
Date:
|
 |
|
10-Jun-07
|
Release date:
|
24-Jul-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P11172
(UMPS_HUMAN) -
Uridine 5'-monophosphate synthase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
480 a.a.
262 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 6 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.2.10
- orotate phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + diphosphate = orotate + 5-phospho-alpha-D-ribose 1-diphosphate
|
 |
 |
 |
 |
 |
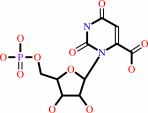 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
diphosphate
Bound ligand (Het Group name = )
matches with 87.50% similarity
|
=
|
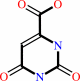 orotate
orotate
|
+
|
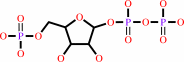 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

