 |
PDBsum entry 1bll

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
1bll
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.11.1
- leucyl aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of an N-terminal amino acid, Xaa-|-Xbb-, in which Xaa is preferably Leu, but may be other amino acids including Pro although not Arg or Lys, and Xbb may be Pro.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.4.11.5
- prolyl aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of a N-terminal proline from a peptide.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.4.13.23
- cysteinylglycine-S-conjugate dipeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an S-substituted L-cysteinylglycine + H2O = an S-substituted L-cysteine + glycine
|
 |
 |
 |
 |
 |
S-substituted L-cysteinylglycine
|
+
|
H2O
|
=
|
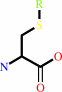 S-substituted L-cysteine
S-substituted L-cysteine
|
+
|
glycine
Bound ligand (Het Group name = )
matches with 55.56% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:8465-8478
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray crystallographic determination of the structure of bovine lens leucine aminopeptidase complexed with amastatin: formulation of a catalytic mechanism featuring a gem-diolate transition state.
|
|
H.Kim,
W.N.Lipscomb.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of the complex of bovine lens leucine aminopeptidase (blLAP) with
the slow-, tight-binding inhibitor amastatin has been determined by X-ray
crystallography. X-ray diffraction data were collected at -150 degrees C from a
single blLAP-amastatin crystal which under the data collection conditions was of
the space group P6(3)22 with unit cell parameters a = 130.3 A and c = 121.9 A.
The structure of the blLAP-amastatin complex was determined by molecular
replacement, using the structure of native blLAP as the starting model.
Refinement of the blLAP-amastatin model plus 132 water molecules against data
from 10.0- to 2.4-A resolution resulted in a final structure with a
crystallographic residual of 0.198. The binding mode of amastatin is similar to
that of bestatin, the structure of whose complex with blLAP has previously been
determined. Of particular note, the N-terminus-to-C-terminus orientation of the
two bound inhibitors is the same. The two N-terminal residues of amastatin and
bestatin occupy the same binding sites, which are most likely S1 and S'1. The
slow binding of amastatin and bestatin may be partially attributable to a
binding mechanism in which the two active site metals are sequentially
coordinated by the P1 amino and hydroxyl groups of these inhibitors. A catalytic
mechanism for blLAP is proposed based on the binding modes of amastatin and
bestatin and plausible binding modes of a dipeptide substrate and its putative
gem-diolate transition state which were modeled into the active site of blLAP
after the binding mode of amastatin. The proposed catalytic mechanism invokes
roles for the catalytic metals in binding and activating the substrate and in
stabilizing the transition state. The mechanism also includes roles for Asp-255
as a general base, Arg-336 as an additional electrophilic substrate activator
and transition state stabilizer, and Lys-262 as a proton shuttle.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Pícha,
R.Liboska,
M.Buděšínský,
J.Jiráček,
M.Pawełczak,
and
A.Mucha
(2011).
Unusual activity pattern of leucine aminopeptidase inhibitors based on phosphorus containing derivatives of methionine and norleucine.
|
| |
J Enzyme Inhib Med Chem,
26,
155-161.
|
 |
|
|
|
|
 |
S.McGowan,
C.A.Oellig,
W.A.Birru,
T.T.Caradoc-Davies,
C.M.Stack,
J.Lowther,
T.Skinner-Adams,
A.Mucha,
P.Kafarski,
J.Grembecka,
K.R.Trenholme,
A.M.Buckle,
D.L.Gardiner,
J.P.Dalton,
and
J.C.Whisstock
(2010).
Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases.
|
| |
Proc Natl Acad Sci U S A,
107,
2449-2454.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.M.McArdle,
and
R.J.Quinn
(2007).
Identification of protein fold topology shared between different folds inhibited by natural products.
|
| |
Chembiochem,
8,
788-798.
|
 |
|
|
|
|
 |
I.Herrera-Camacho,
N.H.Rosas-Murrieta,
A.Rojo-Domínguez,
L.Millán,
J.Reyes-Leyva,
G.Santos-López,
and
P.Suárez-Rendueles
(2007).
Biochemical characterization and structural prediction of a novel cytosolic leucyl aminopeptidase of the M17 family from Schizosaccharomyces pombe.
|
| |
FEBS J,
274,
6228-6240.
|
 |
|
|
|
|
 |
P.J.Mikulecky,
and
A.L.Feig
(2006).
Heat capacity changes associated with nucleic acid folding.
|
| |
Biopolymers,
82,
38-58.
|
 |
|
|
|
|
 |
B.Bennett,
W.E.Antholine,
V.M.D'souza,
G.Chen,
L.Ustinyuk,
and
R.C.Holz
(2002).
Structurally distinct active sites in the copper(II)-substituted aminopeptidases from Aeromonas proteolytica and Escherichia coli.
|
| |
J Am Chem Soc,
124,
13025-13034.
|
 |
|
|
|
|
 |
K.Håkansson,
and
C.G.Miller
(2002).
Structure of peptidase T from Salmonella typhimurium.
|
| |
Eur J Biochem,
269,
443-450.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Erhardt,
and
J.Weston
(2002).
Development of a working model of the active site in bovine lens leucine aminopeptidase: a density functional investigation.
|
| |
Chembiochem,
3,
101-104.
|
 |
|
|
|
|
 |
Y.Q.Gu,
and
L.L.Walling
(2002).
Identification of residues critical for activity of the wound-induced leucine aminopeptidase (LAP-A) of tomato.
|
| |
Eur J Biochem,
269,
1630-1640.
|
 |
|
|
|
|
 |
R.Gilboa,
A.Spungin-Bialik,
G.Wohlfahrt,
D.Schomburg,
S.Blumberg,
and
G.Shoham
(2001).
Interactions of Streptomyces griseus aminopeptidase with amino acid reaction products and their implications toward a catalytic mechanism.
|
| |
Proteins,
44,
490-504.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Schomburg,
H.Kollmus,
S.Friedrichsen,
and
K.Bauer
(2000).
Molecular characterization of a puromycin-insensitive leucyl-specific aminopeptidase, PILS-AP.
|
| |
Eur J Biochem,
267,
3198-3207.
|
 |
|
|
|
|
 |
M.Abramić,
D.Schleuder,
L.Dolovcak,
W.Schröder,
K.Strupat,
D.Sagi,
J.Peter-Katalini,
and
L.Vitale
(2000).
Human and rat dipeptidyl peptidase III: biochemical and mass spectrometric arguments for similarities and differences.
|
| |
Biol Chem,
381,
1233-1243.
|
 |
|
|
|
|
 |
R.Gilboa,
H.M.Greenblatt,
M.Perach,
A.Spungin-Bialik,
U.Lessel,
G.Wohlfahrt,
D.Schomburg,
S.Blumberg,
and
G.Shoham
(2000).
Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
551-558.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Mathew,
T.M.Knox,
and
C.G.Miller
(2000).
Salmonella enterica serovar typhimurium peptidase B is a leucyl aminopeptidase with specificity for acidic amino acids.
|
| |
J Bacteriol,
182,
3383-3393.
|
 |
|
|
|
|
 |
K.M.Huntington,
D.L.Bienvenue,
Y.Wei,
B.Bennett,
R.C.Holz,
and
D.Pei
(1999).
Slow-binding inhibition of the aminopeptidase from Aeromonas proteolytica by peptide thiols: synthesis and spectroscopic characterization.
|
| |
Biochemistry,
38,
15587-15596.
|
 |
|
|
|
|
 |
N.Sträter,
L.Sun,
E.R.Kantrowitz,
and
W.N.Lipscomb
(1999).
A bicarbonate ion as a general base in the mechanism of peptide hydrolysis by dizinc leucine aminopeptidase.
|
| |
Proc Natl Acad Sci U S A,
96,
11151-11155.
|
 |
|
|
|
|
 |
Y.Q.Gu,
F.M.Holzer,
and
L.L.Walling
(1999).
Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of tomato.
|
| |
Eur J Biochem,
263,
726-735.
|
 |
|
|
|
|
 |
G.Chen,
T.Edwards,
V.M.D'souza,
and
R.C.Holz
(1997).
Mechanistic studies on the aminopeptidase from Aeromonas proteolytica: a two-metal ion mechanism for peptide hydrolysis.
|
| |
Biochemistry,
36,
4278-4286.
|
 |
|
|
|
|
 |
B.Chevrier,
H.D'Orchymont,
C.Schalk,
C.Tarnus,
and
D.Moras
(1996).
The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit.
|
| |
Eur J Biochem,
237,
393-398.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Tarnus,
J.M.Rémy,
and
H.d'Orchymont
(1996).
3-Amino-2-hydroxy-propionaldehyde and 3-amino-1-hydroxy-propan-2-one derivatives: new classes of aminopeptidase inhibitors.
|
| |
Bioorg Med Chem,
4,
1287-1297.
|
 |
|
|
|
|
 |
W.L.Mock,
and
Y.Liu
(1995).
Hydrolysis of picolinylprolines by prolidase. A general mechanism for the dual-metal ion containing aminopeptidases.
|
| |
J Biol Chem,
270,
18437-18446.
|
 |
|
|
|
|
 |
B.Chevrier,
C.Schalk,
H.D'Orchymont,
J.M.Rondeau,
D.Moras,
and
C.Tarnus
(1994).
Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family.
|
| |
Structure,
2,
283-291.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

