 |
PDBsum entry 4zqn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/oxidoreductase inhibitor
|
PDB id
|
|

|

|
4zqn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/oxidoreductase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from mycobacterium tuberculosis in the complex with imp and the inhibitor p41
|
|
Structure:
|
 |
Inosine-5'-monophosphate dehydrogenase,inosine-5'- monophosphate dehydrogenase. Chain: a. Fragment: unp residues 1-125 and 253-529 linked by linker (gly gly). Synonym: impdh. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis (strain atcc 25618 / h37rv). Organism_taxid: 83332. Strain: atcc 25618 / h37rv. Gene: guab, guab2, rv3411c, mtcy78.17. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.180
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
Y.Kim,M.Makowska-Grzyska,M.Gu,M.Kavitha,L.Hedstrom,W.F.Anderson, A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
M.Makowska-Grzyska
et al.
(2015).
Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One,
10,
e0138976.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
10-May-15
|
Release date:
|
17-Jun-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WKI7
(IMDH_MYCTU) -
Inosine-5'-monophosphate dehydrogenase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
529 a.a.
353 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.205
- Imp dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + NAD+ + H2O = XMP + NADH + H+
|
 |
 |
 |
 |
 |
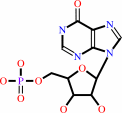 IMP
IMP
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
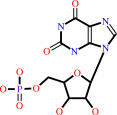 XMP
XMP
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Plos One
10:e0138976
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
|
|
M.Makowska-Grzyska,
Y.Kim,
S.K.Gorla,
Y.Wei,
K.Mandapati,
M.Zhang,
N.Maltseva,
G.Modi,
H.I.Boshoff,
M.Gu,
C.Aldrich,
G.D.Cuny,
L.Hedstrom,
A.Joachimiak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tuberculosis (TB) remains a worldwide problem and the need for new drugs is
increasingly more urgent with the emergence of multidrug- and extensively-drug
resistant TB. Inosine 5'-monophosphate dehydrogenase 2 (IMPDH2) from
Mycobacterium tuberculosis (Mtb) is an attractive drug target. The enzyme
catalyzes the conversion of inosine 5'-monophosphate into xanthosine
5'-monophosphate with the concomitant reduction of NAD+ to NADH. This reaction
controls flux into the guanine nucleotide pool. We report seventeen selective
IMPDH inhibitors with antitubercular activity. The crystal structures of a
deletion mutant of MtbIMPDH2 in the apo form and in complex with the product XMP
and substrate NAD+ are determined. We also report the structures of complexes
with IMP and three structurally distinct inhibitors, including two with
antitubercular activity. These structures will greatly facilitate the
development of MtbIMPDH2-targeted antibiotics.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

