 |
PDBsum entry 3ecm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.53
- 3',5'-cyclic-AMP phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3',5'-cyclic AMP + H2O = AMP + H+
|
 |
 |
 |
 |
 |
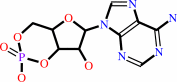 3',5'-cyclic AMP
3',5'-cyclic AMP
|
+
|
H2O
|
=
|
 AMP
AMP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
47:12760-12768
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity.
|
|
H.Wang,
Z.Yan,
S.Yang,
J.Cai,
H.Robinson,
H.Ke.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cyclic nucleotide phosphodiesterase-8 (PDE8) is a family of cAMP-specific
enzymes and plays important roles in many biological processes, including T-cell
activation, testosterone production, adrenocortical hyperplasia, and thyroid
function. However, no PDE8 selective inhibitors are available for trial
treatment of human diseases. Here we report kinetic properties of the highly
active PDE8A1 catalytic domain prepared from refolding and its crystal
structures in the unliganded and 3-isobutyl-1-methylxanthine (IBMX) bound forms
at 1.9 and 2.1 A resolutions, respectively. The PDE8A1 catalytic domain has a
K(M) of 1.8 microM, V(max) of 6.1 micromol/min/mg, a k(cat) of 4.0 s(-1) for
cAMP, and a K(M) of 1.6 mM, V(max) of 2.5 micromol/min/mg, a k(cat) of 1.6 s(-1)
for cGMP, thus indicating that the substrate specificity of PDE8 is dominated by
K(M). The structure of the PDE8A1 catalytic domain has similar topology as those
of other PDE families but contains two extra helices around Asn685-Thr710. Since
this fragment is distant from the active site of the enzyme, its impact on the
catalysis is unclear. The PDE8A1 catalytic domain is insensitive to the IBMX
inhibition (IC(50) = 700 microM). The unfavorable interaction of IBMX in the
PDE8A1-IBMX structure suggests an important role of Tyr748 in the inhibitor
binding. Indeed, the mutation of Tyr748 to phenylalanine increases the PDE8A1
sensitivity to several nonselective or family selective PDE inhibitors. Thus,
the structural and mutagenesis studies provide not only insight into the
enzymatic properties but also guidelines for design of PDE8 selective inhibitors.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Wang,
X.Luo,
M.Ye,
J.Hou,
H.Robinson,
and
H.Ke
(2010).
Insight into binding of phosphodiesterase-9A selective inhibitors by crystal structures and mutagenesis.
|
| |
J Med Chem,
53,
1726-1731.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Weeks,
J.D.Corbin,
and
S.H.Francis
(2009).
Interactions between cyclic nucleotide phosphodiesterase 11 catalytic site and substrates or tadalafil and role of a critical Gln-869 hydrogen bond.
|
| |
J Pharmacol Exp Ther,
331,
133-141.
|
 |
|
|
|
|
 |
J.Pandit,
M.D.Forman,
K.F.Fennell,
K.S.Dillman,
and
F.S.Menniti
(2009).
Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
|
| |
Proc Natl Acad Sci U S A,
106,
18225-18230.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

