 |
PDBsum entry 2plc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.6.1.13
- phosphatidylinositol diacylglycerol-lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
1-Phosphatidyl-myo-inositol Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) = 1D-myo-inositol 1,2-cyclic phosphate + a 1,2-diacyl-sn-glycerol
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol)
|
=
|
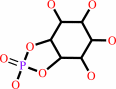 1D-myo-inositol 1,2-cyclic phosphate
1D-myo-inositol 1,2-cyclic phosphate
|
+
|
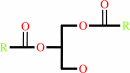 1,2-diacyl-sn-glycerol
1,2-diacyl-sn-glycerol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
273:269-282
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the phosphatidylinositol-specific phospholipase C from the human pathogen Listeria monocytogenes.
|
|
J.Moser,
B.Gerstel,
J.E.Meyer,
T.Chakraborty,
J.Wehland,
D.W.Heinz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray crystal structure of the phosphatidylinositol-specific phospholipase C
(PI-PLC) from the human pathogen Listeria monocytogenes has been determined both
in free form at 2.0 A resolution, and in complex with the competitive inhibitor
myo-inositol at 2.6 A resolution. The structure was solved by a combination of
molecular replacement using the structure of Bacillus cereus PI-PLC and single
isomorphous replacement. The enzyme consists of a single (beta alpha)8-barrel
domain with the active site located at the C-terminal side of the beta-barrel.
Unlike other (beta alpha)8-barrels, the barrel in PI-PLC is open because it
lacks hydrogen bonding interactions between beta-strands V and VI. myo-Inositol
binds to the active site pocket by making specific hydrogen bonding interactions
with a number of charged amino acid side-chains as well as a coplanar stacking
interaction with a tyrosine residue. Despite a relatively low sequence identity
of approximately 24%, the structure is highly homologous to that of B.cereus
PI-PLC with an r.m.s. deviation for 228 common C alpha positions of 1.46 A.
Larger differences are found for loop regions that accommodate most of the
numerous amino acid insertions and deletions. The active site pocket is also
well conserved with only two amino acid replacements directly implicated in
inositol binding.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Reaction catalyzed by L.monocytogenes PI-PLC.
|
 |
Figure 10.
Figure 10. Side-by-side view of the molecular surfaces of BPI-PLC (left) and LPI-PLC (right) showing the C-terminal
side of the b-barrel with the active site pocket in the center, where negative potentials on the surface are shaded in
red and positive potentials in blue. myo-Inositol bound to the active site pocket is depicted by yellow bonds. The
hydrophobic ridge is designated by A, the extended GPI-binding site by B. The picture was generated using the pro-
gram GRASP (Nicholls, 1993).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(1997,
273,
269-282)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Böttcher,
and
S.A.Sieber
(2009).
Beta-lactones decrease the intracellular virulence of Listeria monocytogenes in macrophages.
|
| |
ChemMedChem,
4,
1260-1263.
|
 |
|
|
|
|
 |
W.Chen,
H.Goldfine,
B.Ananthanarayanan,
W.Cho,
and
M.F.Roberts
(2009).
Listeria monocytogenes phosphatidylinositol-specific phospholipase C: Kinetic activation and homing in on different interfaces.
|
| |
Biochemistry,
48,
3578-3592.
|
 |
|
|
|
|
 |
A.S.Ozyurt,
and
T.L.Selby
(2008).
Computational active site analysis of molecular pathways to improve functional classification of enzymes.
|
| |
Proteins,
72,
184-196.
|
 |
|
|
|
|
 |
S.Témoin,
S.M.Roche,
O.Grépinet,
Y.Fardini,
and
P.Velge
(2008).
Multiple point mutations in virulence genes explain the low virulence of Listeria monocytogenes field strains.
|
| |
Microbiology,
154,
939-948.
|
 |
|
|
|
|
 |
C.Shao,
X.Shi,
H.Wehbi,
C.Zambonelli,
J.F.Head,
B.A.Seaton,
and
M.F.Roberts
(2007).
Dimer structure of an interfacially impaired phosphatidylinositol-specific phospholipase C.
|
| |
J Biol Chem,
282,
9228-9235.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Wei,
L.A.Zenewicz,
and
H.Goldfine
(2005).
Listeria monocytogenes phosphatidylinositol-specific phospholipase C has evolved for virulence by greatly reduced activity on GPI anchors.
|
| |
Proc Natl Acad Sci U S A,
102,
12927-12931.
|
 |
|
|
|
|
 |
Z.Wei,
P.Schnupf,
M.A.Poussin,
L.A.Zenewicz,
H.Shen,
and
H.Goldfine
(2005).
Characterization of Listeria monocytogenes expressing anthrolysin O and phosphatidylinositol-specific phospholipase C from Bacillus anthracis.
|
| |
Infect Immun,
73,
6639-6646.
|
 |
|
|
|
|
 |
J.Feng,
W.D.Bradley,
and
M.F.Roberts
(2003).
Optimizing the interfacial binding and activity of a bacterial phosphatidylinositol-specific phospholipase C.
|
| |
J Biol Chem,
278,
24651-24657.
|
 |
|
|
|
|
 |
F.J.Sharom,
and
M.T.Lehto
(2002).
Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C.
|
| |
Biochem Cell Biol,
80,
535-549.
|
 |
|
|
|
|
 |
J.Feng,
H.Wehbi,
and
M.F.Roberts
(2002).
Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C.
|
| |
J Biol Chem,
277,
19867-19875.
|
 |
|
|
|
|
 |
R.J.Kubiak,
X.Yue,
R.J.Hondal,
C.Mihai,
M.D.Tsai,
and
K.S.Bruzik
(2001).
Involvement of the Arg-Asp-His catalytic triad in enzymatic cleavage of the phosphodiester bond.
|
| |
Biochemistry,
40,
5422-5432.
|
 |
|
|
|
|
 |
H.Goldfine,
S.J.Wadsworth,
and
N.C.Johnston
(2000).
Activation of host phospholipases C and D in macrophages after infection with Listeria monocytogenes.
|
| |
Infect Immun,
68,
5735-5741.
|
 |
|
|
|
|
 |
T.Bannam,
and
H.Goldfine
(1999).
Mutagenesis of active-site histidines of Listeria monocytogenes phosphatidylinositol-specific phospholipase C: effects on enzyme activity and biological function.
|
| |
Infect Immun,
67,
182-186.
|
 |
|
|
|
|
 |
M.Katan
(1998).
Families of phosphoinositide-specific phospholipase C: structure and function.
|
| |
Biochim Biophys Acta,
1436,
5.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

