 |
PDBsum entry 2o8h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.17
- 3',5'-cyclic-nucleotide phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H+
|
 |
 |
 |
 |
 |
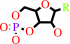 nucleoside 3',5'-cyclic phosphate
nucleoside 3',5'-cyclic phosphate
|
+
|
H2O
|
=
|
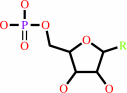 nucleoside 5'-phosphate
nucleoside 5'-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
50:182-185
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Discovery of a series of 6,7-dimethoxy-4-pyrrolidylquinazoline PDE10A inhibitors.
|
|
T.A.Chappie,
J.M.Humphrey,
M.P.Allen,
K.G.Estep,
C.B.Fox,
L.A.Lebel,
S.Liras,
E.S.Marr,
F.S.Menniti,
J.Pandit,
C.J.Schmidt,
M.Tu,
R.D.Williams,
F.V.Yang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A papaverine based pharmacophore model for PDE10A inhibition was generated via
SBDD and used to design a library of 4-amino-6,7-dimethoxyquinazolines. From
this library emerged an aryl ether pyrrolidyl 6,7-dimethoxyquinazoline series
that became the focal point for additional modeling, X-ray, and synthetic
efforts toward increasing PDE10A inhibitory potency and selectivity versus
PDE3A/B. These efforts culminated in the discovery of 29, a potent and selective
brain penetrable inhibitor of PDE10A.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Duplais,
A.Krasovskiy,
A.Wattenberg,
and
B.H.Lipshutz
(2010).
Cross-couplings between benzylic and aryl halides "on water": synthesis of diarylmethanes.
|
| |
Chem Commun (Camb),
46,
562-564.
|
 |
|
|
|
|
 |
O.A.Andersen,
D.L.Schönfeld,
I.Toogood-Johnson,
B.Felicetti,
C.Albrecht,
T.Fryatt,
M.Whittaker,
D.Hallett,
and
J.Barker
(2009).
Cross-linking of protein crystals as an aid in the generation of binary protein-ligand crystal complexes, exemplified by the human PDE10a-papaverine structure.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
872-874.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Siuciak
(2008).
The role of phosphodiesterases in schizophrenia : therapeutic implications.
|
| |
CNS Drugs,
22,
983-993.
|
 |
|
|
|
|
 |
M.Amatore,
and
C.Gosmini
(2008).
Synthesis of functionalised diarylmethanes via a cobalt-catalysed cross-coupling of arylzinc species with benzyl chlorides.
|
| |
Chem Commun (Camb),
(),
5019-5021.
|
 |
|
|
|
|
 |
S.E.Martinez,
C.C.Heikaus,
R.E.Klevit,
and
J.A.Beavo
(2008).
The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change.
|
| |
J Biol Chem,
283,
25913-25919.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

