 |
PDBsum entry 2isl

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Flavoprotein
|
 |
|
Title:
|
 |
Blub bound to reduced flavin (fmnh2) and molecular oxygen. (Clear crystal form)
|
|
Structure:
|
 |
Blub. Chain: a, b, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Sinorhizobium meliloti. Organism_taxid: 382. Gene: blub. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.90Å
|
R-factor:
|
0.217
|
R-free:
|
0.285
|
|
|
Authors:
|
 |
N.A.Larsen,M.E.Taga,A.R.Howard-Jones,C.T.Walsh,G.C.Walker
|
Key ref:

|
 |
M.E.Taga
et al.
(2007).
BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature,
446,
449-453.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Oct-06
|
Release date:
|
27-Mar-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q92PC8
(BLUB_RHIME) -
5,6-dimethylbenzimidazole synthase from Rhizobium meliloti (strain 1021)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
227 a.a.
219 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.79
- aerobic 5,6-dimethylbenzimidazole synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
FMNH2 + O2 = dialurate + 5,6-dimethylbenzimidazole + D-erythrose 4-phosphate + H+
|
 |
 |
 |
 |
 |
FMNH2
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
O2
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
dialurate
|
+
|
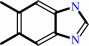 5,6-dimethylbenzimidazole
5,6-dimethylbenzimidazole
|
+
|
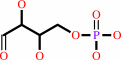 D-erythrose 4-phosphate
D-erythrose 4-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
446:449-453
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
BluB cannibalizes flavin to form the lower ligand of vitamin B12.
|
|
M.E.Taga,
N.A.Larsen,
A.R.Howard-Jones,
C.T.Walsh,
G.C.Walker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Vitamin B12 (cobalamin) is among the largest known non-polymeric natural
products, and the only vitamin synthesized exclusively by microorganisms. The
biosynthesis of the lower ligand of vitamin B(12), 5,6-dimethylbenzimidazole
(DMB), is poorly understood. Recently, we discovered that a Sinorhizobium
meliloti gene, bluB, is necessary for DMB biosynthesis. Here we show that BluB
triggers the unprecedented fragmentation and contraction of the bound flavin
mononucleotide cofactor and cleavage of the ribityl tail to form DMB and
D-erythrose 4-phosphate. Our structural analysis shows that BluB resembles an
NAD(P)H-flavin oxidoreductase, except that its unusually tight binding pocket
accommodates flavin mononucleotide but not NAD(P)H. We characterize
crystallographically an early intermediate along the reaction coordinate,
revealing molecular oxygen poised over reduced flavin. Thus, BluB isolates and
directs reduced flavin to activate molecular oxygen for its own cannibalization.
This investigation of the biosynthesis of DMB provides clarification of an
aspect of vitamin B12 that was otherwise incomplete, and may contribute to a
better understanding of vitamin B12-related disease.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2: Structure of BluB. a, Ribbon diagram of BluB
with FMN in the binding pocket (stick representation). The
two-fold axis is perpendicular to the plane of the figure. b, E.
coli nitroreductase NfsB (Protein Data Bank 1ICR)^21 with FMN
and nicotinic acid (stick representation) in the binding pocket.
c, Cross-section of BluB's molecular surface. The two-fold axis
lies along the y axis such that the si-face of FMN is viewed on
the left and re-face on the right. The surface is coloured
according to electrostatic potential, where blue is
electropositive, red is electronegative and k[B] is Boltzmann's
constant. The back and front of the surface are cut away to
reveal the FMN binding pocket buried in the dimer interface. The
pocket wraps snugly around FMN, preventing interaction with
other substrates. d, The BluB ribbon diagram has been coloured
according to B-factor. Red represents flexible regions that may
control or gate access to the active site.
|
 |
Figure 3.
Figure 3: Active site of BluB. a, b, The active site with
oxidized FMN (a) and reduced FMN (b), viewed from the re-face.
H-bonds are represented as dashed lines. For clarity, water
molecules are not rendered in this view. The rearrangement in
H-bonds around N1 reflects the change in protonation in the
reduced structure. Asp 32 may also form a close contact with C1'
of the ribityl chain, suggesting a potential catalytic role for
this residue. c, d, Side views of the active site in the
oxidized (c) and reduced (d) structures. For clarity, Arg 34 has
not been rendered in this view. The sigma-A weighted 2F[o]-F[c]
electron density map is contoured at 1  and
coloured grey (around protein side chains), blue (around
FMN/FMNH[2]) and red (around water/oxygen) to enhance contrast. and
coloured grey (around protein side chains), blue (around
FMN/FMNH[2]) and red (around water/oxygen) to enhance contrast.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(2007,
446,
449-453)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Ortiz-Guerrero,
M.C.Polanco,
F.J.Murillo,
S.Padmanabhan,
and
M.Elías-Arnanz
(2011).
Light-dependent gene regulation by a coenzyme B12-based photoreceptor.
|
| |
Proc Natl Acad Sci U S A,
108,
7565-7570.
|
 |
|
|
|
|
 |
A.M.Mowafy,
T.Kurihara,
A.Kurata,
T.Uemura,
and
N.Esaki
(2010).
2-haloacrylate hydratase, a new class of flavoenzyme that catalyzes the addition of water to the substrate for dehalogenation.
|
| |
Appl Environ Microbiol,
76,
6032-6037.
|
 |
|
|
|
|
 |
C.C.Fowler,
E.D.Brown,
and
Y.Li
(2010).
Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli.
|
| |
Chem Biol,
17,
756-765.
|
 |
|
|
|
|
 |
H.R.Bonomi,
M.I.Marchesini,
S.Klinke,
J.E.Ugalde,
V.Zylberman,
R.A.Ugalde,
D.J.Comerci,
and
F.A.Goldbaum
(2010).
An atypical riboflavin pathway is essential for Brucella abortus virulence.
|
| |
PLoS One,
5,
e9435.
|
 |
|
|
|
|
 |
M.J.Gray,
and
J.C.Escalante-Semerena
(2010).
A new pathway for the synthesis of α-ribazole-phosphate in Listeria innocua.
|
| |
Mol Microbiol,
77,
1429-1438.
|
 |
|
|
|
|
 |
M.Vujkovic,
E.A.Steegers,
J.van Meurs,
N.Yazdanpanah,
I.A.van Rooij,
A.G.Uitterlinden,
and
R.P.Steegers-Theunissen
(2010).
The maternal homocysteine pathway is influenced by riboflavin intake and MTHFR polymorphisms without affecting the risk of orofacial clefts in the offspring.
|
| |
Eur J Clin Nutr,
64,
266-273.
|
 |
|
|
|
|
 |
C.T.Jurgenson,
T.P.Begley,
and
S.E.Ealick
(2009).
The structural and biochemical foundations of thiamin biosynthesis.
|
| |
Annu Rev Biochem,
78,
569-603.
|
 |
|
|
|
|
 |
M.Grininger,
H.Staudt,
P.Johansson,
J.Wachtveitl,
and
D.Oesterhelt
(2009).
Dodecin is the key player in flavin homeostasis of archaea.
|
| |
J Biol Chem,
284,
13068-13076.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Gray,
and
J.C.Escalante-Semerena
(2009).
In vivo analysis of cobinamide salvaging in Rhodobacter sphaeroides strain 2.4.1.
|
| |
J Bacteriol,
191,
3842-3851.
|
 |
|
|
|
|
 |
M.J.Gray,
and
J.C.Escalante-Semerena
(2009).
The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: a critical activity of the cobamide remodelling system of Rhodobacter sphaeroides.
|
| |
Mol Microbiol,
74,
1198-1210.
|
 |
|
|
|
|
 |
P.F.Widboom,
and
S.D.Bruner
(2009).
Complex oxidation chemistry in the biosynthetic pathways to vancomycin/teicoplanin antibiotics.
|
| |
Chembiochem,
10,
1757-1764.
|
 |
|
|
|
|
 |
S.R.Thomas,
P.M.McTamney,
J.M.Adler,
N.Laronde-Leblanc,
and
S.E.Rokita
(2009).
Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
|
| |
J Biol Chem,
284,
19659-19667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Pérez-Reinado,
M.D.Roldán,
F.Castillo,
and
C.Moreno-Vivián
(2008).
The NprA nitroreductase required for 2,4-dinitrophenol reduction in Rhodobacter capsulatus is a dihydropteridine reductase.
|
| |
Environ Microbiol,
10,
3174-3183.
|
 |
|
|
|
|
 |
K.E.Gibson,
H.Kobayashi,
and
G.C.Walker
(2008).
Molecular determinants of a symbiotic chronic infection.
|
| |
Annu Rev Genet,
42,
413-441.
|
 |
|
|
|
|
 |
L.L.Grochowski,
and
R.H.White
(2008).
Promiscuous anaerobes: new and unconventional metabolism in methanogenic archaea.
|
| |
Ann N Y Acad Sci,
1125,
190-214.
|
 |
|
|
|
|
 |
M.D.Roldán,
E.Pérez-Reinado,
F.Castillo,
and
C.Moreno-Vivián
(2008).
Reduction of polynitroaromatic compounds: the bacterial nitroreductases.
|
| |
FEMS Microbiol Rev,
32,
474-500.
|
 |
|
|
|
|
 |
M.E.Taga,
and
G.C.Walker
(2008).
Pseudo-B12 joins the cofactor family.
|
| |
J Bacteriol,
190,
1157-1159.
|
 |
|
|
|
|
 |
P.J.Anderson,
J.Lango,
C.Carkeet,
A.Britten,
B.Kräutler,
B.D.Hammock,
and
J.R.Roth
(2008).
One pathway can incorporate either adenine or dimethylbenzimidazole as an alpha-axial ligand of B12 cofactors in Salmonella enterica.
|
| |
J Bacteriol,
190,
1160-1171.
|
 |
|
|
|
|
 |
F.Rébeillé,
S.Ravanel,
A.Marquet,
R.R.Mendel,
M.E.Webb,
A.G.Smith,
and
M.J.Warren
(2007).
Roles of vitamins B5, B8, B9, B12 and molybdenum cofactor at cellular and organismal levels.
|
| |
Nat Prod Rep,
24,
949-962.
|
 |
|
|
|
|
 |
J.C.Escalante-Semerena
(2007).
Conversion of cobinamide into adenosylcobamide in bacteria and archaea.
|
| |
J Bacteriol,
189,
4555-4560.
|
 |
|
|
|
|
 |
J.W.Choi,
J.Lee,
N.Kosuke,
C.H.Jung,
and
J.S.Kim
(2007).
Crystallization and preliminary X-ray diffraction analysis of ydjA, a minimal nitroreductase from Escherichia coli K12.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
1064-1066.
|
 |
|
|
|
|
 |
S.E.Ealick,
and
T.P.Begley
(2007).
Biochemistry: molecular cannibalism.
|
| |
Nature,
446,
387-388.
|
 |
|
|
|
|
 |
S.Fetzner
(2007).
Cofactor-independent oxygenases go it alone.
|
| |
Nat Chem Biol,
3,
374-375.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

