 |
PDBsum entry 2cdc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2cdc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Sulfolobus solfataricus glucose dehydrogenase 1 in complex with NADP and xylose
|
|
Structure:
|
 |
Glucose dehydrogenase glucose 1-dehydrogenase, dhg-1. Chain: a, b, c, d. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Sulfolobus solfataricus. Organism_taxid: 2287. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.206
|
R-free:
|
0.239
|
|
|
Authors:
|
 |
C.C.Milburn,H.J.Lamble,A.Theodossis,D.W.Hough,M.J.Danson,G.L.Taylor
|
Key ref:

|
 |
C.C.Milburn
et al.
(2006).
The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J Biol Chem,
281,
14796-14804.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
23-Jan-06
|
Release date:
|
22-Mar-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O93715
(GLCDH_SACSO) -
Glucose 1-dehydrogenase from Saccharolobus solfataricus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
366 a.a.
359 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.120
- galactose 1-dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-galactose + NADP+ = D-galactono-1,5-lactone + NADPH + H+
|
 |
 |
 |
 |
 |
D-galactose
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
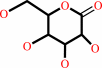 D-galactono-1,5-lactone
D-galactono-1,5-lactone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.359
- aldose 1-dehydrogenase [NAD(P)(+)].
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an aldopyranose + NADP+ = aldono-1,5-lactone + NADPH + H+
|
|
2.
|
an aldopyranose + NAD+ = aldono-1,5-lactone + NADH + H+
|
|
 |
 |
 |
 |
 |
aldopyranose
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
aldono-1,5-lactone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
aldopyranose
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
aldono-1,5-lactone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.47
- glucose 1-dehydrogenase [NAD(P)(+)].
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
D-glucose + NADP+ = D-glucono-1,5-lactone + NADPH + H+
|
|
2.
|
D-glucose + NAD+ = D-glucono-1,5-lactone + NADH + H+
|
|
 |
 |
 |
 |
 |
D-glucose
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 D-glucono-1,5-lactone
D-glucono-1,5-lactone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
D-glucose
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 D-glucono-1,5-lactone
D-glucono-1,5-lactone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.48
- D-galactose 1-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-galactose + NAD+ = D-galactono-1,4-lactone + NADH + H+
|
 |
 |
 |
 |
 |
D-galactose
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
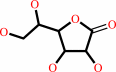 D-galactono-1,4-lactone
D-galactono-1,4-lactone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:14796-14804
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
|
|
C.C.Milburn,
H.J.Lamble,
A.Theodossis,
S.D.Bull,
D.W.Hough,
M.J.Danson,
G.L.Taylor.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The hyperthermophilic archaeon Sulfolobus solfataricus grows optimally above 80
degrees C and utilizes an unusual, promiscuous, non-phosphorylative
Entner-Doudoroff pathway to metabolize both glucose and galactose. The first
enzyme in this pathway, glucose dehydrogenase, catalyzes the oxidation of
glucose to gluconate, but has been shown to have activity with a broad range of
sugar substrates, including glucose, galactose, xylose, and L-arabinose, with a
requirement for the glucose stereo configuration at the C2 and C3 positions.
Here we report the crystal structure of the apo form of glucose dehydrogenase to
a resolution of 1.8 A and a complex with its required cofactor, NADP+, to a
resolution of 2.3 A. A T41A mutation was engineered to enable the trapping of
substrate in the crystal. Complexes of the enzyme with D-glucose and D-xylose
are presented to resolutions of 1.6 and 1.5 A, respectively, that provide
evidence of selectivity for the beta-anomeric, pyranose form of the substrate,
and indicate that this is the productive substrate form. The nature of the
promiscuity of glucose dehydrogenase is also elucidated, and a physiological
role for this enzyme in xylose metabolism is suggested. Finally, the structure
suggests that the mechanism of sugar oxidation by this enzyme may be similar to
that described for human sorbitol dehydrogenase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. Stereo images of the apo SsGDH tetramer (A) and
monomer (B). The A-monomer is shown with the nucleotide-binding
domain in red and the catalytic domain in blue. The position of
the GXGXXG motif is highlighted in yellow. Zinc ions are shown
as magenta spheres, and the catalytic zinc-coordinated water is
shown as a green sphere.In B, the N and C termini of the monomer
are indicated by green and red spheres, respectively.
|
 |
Figure 3.
FIGURE 3. A, glucose bound to the SsGDH active site in the
A-monomer. Coloring is as in Fig. 2B with the mutation T41A
highlighted by orange carbons and the glucose molecule shown
with purple carbons and red oxygens, with both C6-hydroxyl
conformations. Unbiased F[c] -F[c] electron density for the
substrate is shown as green mesh (contoured at 2.25  ).
Hydrogen bonds between the protein and glucose are shown as
broken black lines, and gray broken lines indicate interactions
of 3.5-3.7 Å that are possible hydrogen bonds at the
moment of catalysis. Asp^154 sits below the sugar ring
interacting with the C2- and C3-hydroxyls. B, xylose bound to
the SsGDH active site of monomer A. Coloring is as in A, but the
glucose molecule is shown in the equatorial ).
Hydrogen bonds between the protein and glucose are shown as
broken black lines, and gray broken lines indicate interactions
of 3.5-3.7 Å that are possible hydrogen bonds at the
moment of catalysis. Asp^154 sits below the sugar ring
interacting with the C2- and C3-hydroxyls. B, xylose bound to
the SsGDH active site of monomer A. Coloring is as in A, but the
glucose molecule is shown in the equatorial  -form with purple
carbons and red oxygens, and in the axial ( -form with purple
carbons and red oxygens, and in the axial (  -form) with
wheat-colored carbons. Unbiased F[c] - F[c] electron density for
the substrate is shown as green mesh (contoured at 2.25 -form) with
wheat-colored carbons. Unbiased F[c] - F[c] electron density for
the substrate is shown as green mesh (contoured at 2.25  ).
Hydrogen bonds between the protein and ).
Hydrogen bonds between the protein and  -xylose are shown as
broken black lines, and gray broken lines indicate interactions
of <3.5-3.7 Å that are possible hydrogen bonds at the
moment of catalysis. Hydrogen bonds to the -xylose are shown as
broken black lines, and gray broken lines indicate interactions
of <3.5-3.7 Å that are possible hydrogen bonds at the
moment of catalysis. Hydrogen bonds to the  -form are not shown,
because most, with the exception of the C1-OH interactions, are
maintained and no new hydrogen bonds are formed in the -form are not shown,
because most, with the exception of the C1-OH interactions, are
maintained and no new hydrogen bonds are formed in the  -form.
C, superposition of glucose (green) and xylose (blue) in the
active site of the A-monomer. The two positions for O6 of
glucose are displayed, as are the two positions of O1 of xylose.
Glu^114 undergoes a conformational change between the glucose
and xylose complex structures; the alternative position for this
residue in the xylose structure is depicted in wheat-colored
carbons. -form.
C, superposition of glucose (green) and xylose (blue) in the
active site of the A-monomer. The two positions for O6 of
glucose are displayed, as are the two positions of O1 of xylose.
Glu^114 undergoes a conformational change between the glucose
and xylose complex structures; the alternative position for this
residue in the xylose structure is depicted in wheat-colored
carbons.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
14796-14804)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.Haferkamp,
S.Kutschki,
J.Treichel,
H.Hemeda,
K.Sewczyk,
D.Hoffmann,
M.Zaparty,
and
B.Siebers
(2011).
An additional glucose dehydrogenase from Sulfolobus solfataricus: fine-tuning of sugar degradation?
|
| |
Biochem Soc Trans,
39,
77-81.
|
 |
|
|
|
|
 |
P.J.Baker,
K.L.Britton,
M.Fisher,
J.Esclapez,
C.Pire,
M.J.Bonete,
J.Ferrer,
and
D.W.Rice
(2009).
Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
|
| |
Proc Natl Acad Sci U S A,
106,
779-784.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Potter,
M.Kerou,
H.J.Lamble,
S.D.Bull,
D.W.Hough,
M.J.Danson,
and
G.L.Taylor
(2008).
The structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
1283-1287.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.J.Ettema,
H.Ahmed,
A.C.Geerling,
J.van der Oost,
and
B.Siebers
(2008).
The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) of Sulfolobus solfataricus: a key-enzyme of the semi-phosphorylative branch of the Entner-Doudoroff pathway.
|
| |
Extremophiles,
12,
75-88.
|
 |
|
|
|
|
 |
D.Kehrer,
H.Ahmed,
H.Brinkmann,
and
B.Siebers
(2007).
Glycerate kinase of the hyperthermophilic archaeon Thermoproteus tenax: new insights into the phylogenetic distribution and physiological role of members of the three different glycerate kinase classes.
|
| |
BMC Genomics,
8,
301.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

