 |
PDBsum entry 2c6h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2c6h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of yc-17-bound cytochrome p450 pikc (cyp107l1)
|
|
Structure:
|
 |
Cytochrome p450 monooxygenase. Chain: a, b. Synonym: cytochrome p450 pikc cyp107l1, cytochrome p450 hydroxylase pikc. Engineered: yes
|
|
Source:
|
 |
Streptomyces venezuelae. Organism_taxid: 54571. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.35Å
|
R-factor:
|
0.195
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
D.H.Sherman,S.Li,L.V.Yermalitskaya,Y.Kim,J.A.Smith,M.R.Waterman, L.M.Podust
|
Key ref:

|
 |
D.H.Sherman
et al.
(2006).
The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae.
J Biol Chem,
281,
26289-26297.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Nov-05
|
Release date:
|
03-Jul-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O87605
(PIKC_STRVZ) -
Cytochrome P450 monooxygenase PikC from Streptomyces venezuelae
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
416 a.a.
394 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.15.33
- pikromycin synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
narbomycin + 2 reduced [2Fe-2S]-[ferredoxin] + O2 + 2 H+ = pikromycin + 2 oxidized [2Fe-2S]-[ferredoxin] + H2O
|
|
2.
|
narbomycin + 2 reduced [2Fe-2S]-[ferredoxin] + O2 + 2 H+ = neopikromycin + 2 oxidized [2Fe-2S]-[ferredoxin] + H2O
|
|
3.
|
narbomycin + 4 reduced [2Fe-2S]-[ferredoxin] + 2 O2 + 4 H+ = novapikromycin + 4 oxidized [2Fe-2S]-[ferredoxin] + 2 H2O
|
|
4.
|
10-deoxymethymycin + 2 reduced [2Fe-2S]-[ferredoxin] + O2 + 2 H+ = methymycin + 2 oxidized [2Fe-2S]-[ferredoxin] + H2O
|
|
5.
|
10-deoxymethymycin + 2 reduced [2Fe-2S]-[ferredoxin] + O2 + 2 H+ = neomethymycin + 2 oxidized [2Fe-2S]-[ferredoxin] + H2O
|
|
6.
|
10-deoxymethymycin +
|
|
 |
 |
 |
 |
 |
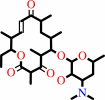 narbomycin
narbomycin
|
+
|
2
×
reduced [2Fe-2S]-[ferredoxin]
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
 O2
O2
|
+
|
2
×
H(+)
|
=
|
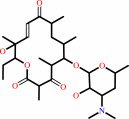 pikromycin
pikromycin
|
+
|
2
×
oxidized [2Fe-2S]-[ferredoxin]
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 narbomycin
narbomycin
|
+
|
2
×
reduced [2Fe-2S]-[ferredoxin]
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
 O2
O2
|
+
|
2
×
H(+)
|
=
|
neopikromycin
|
+
|
2
×
oxidized [2Fe-2S]-[ferredoxin]
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 narbomycin
narbomycin
|
+
|
4
×
reduced [2Fe-2S]-[ferredoxin]
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
 2
×
O2
2
×
O2
|
+
|
4
×
H(+)
|
=
|
novapikromycin
|
+
|
4
×
oxidized [2Fe-2S]-[ferredoxin]
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
10-deoxymethymycin
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
2
×
reduced [2Fe-2S]-[ferredoxin]
|
+
|
 O2
O2
|
+
|
2
×
H(+)
|
=
|
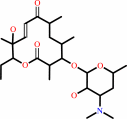 methymycin
methymycin
|
+
|
2
×
oxidized [2Fe-2S]-[ferredoxin]
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
10-deoxymethymycin
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
2
×
reduced [2Fe-2S]-[ferredoxin]
|
+
|
 O2
O2
|
+
|
2
×
H(+)
|
=
|
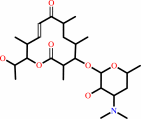 neomethymycin
neomethymycin
|
+
|
2
×
oxidized [2Fe-2S]-[ferredoxin]
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
10-deoxymethymycin
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme-thiolate
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:26289-26297
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae.
|
|
D.H.Sherman,
S.Li,
L.V.Yermalitskaya,
Y.Kim,
J.A.Smith,
M.R.Waterman,
L.M.Podust.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The pikromycin (Pik)/methymycin biosynthetic pathway of Streptomyces venezuelae
represents a valuable system for dissecting the fundamental mechanisms of
modular polyketide biosynthesis, aminodeoxysugar assembly, glycosyltransfer, and
hydroxylation leading to the production of a series of macrolide antibiotics,
including the natural ketolides narbomycin and pikromycin. In this study, we
describe four x-ray crystal structures and allied functional studies for PikC,
the remarkable P450 monooxygenase responsible for production of a number of
related macrolide products from the Pik pathway. The results provide important
new insights into the structural basis for the C10/C12 and C12/C14 hydroxylation
patterns for the 12-(YC-17) and 14-membered ring (narbomycin) macrolides,
respectively. This includes two different ligand-free structures in an
asymmetric unit (resolution 2.1 A) and two co-crystal structures with bound
endogenous substrates YC-17 (resolution 2.35 A)or narbomycin (resolution 1.7 A).
A central feature of the enzyme-substrate interaction involves anchoring of the
desosamine residue in two alternative binding pockets based on a series of
distinct amino acid residues that form a salt bridge and a hydrogen-bonding
network with the deoxysugar C3' dimethylamino group. Functional significance of
the salt bridge was corroborated by site-directed mutagenesis that revealed a
key role for Glu-94 in YC-17 binding and Glu-85 for narbomycin binding. Taken
together, the x-ray structure analysis, site-directed mutagenesis, and
corresponding product distribution studies reveal that PikC substrate tolerance
and product diversity result from a combination of alternative anchoring modes
rather than an induced fit mechanism.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIGURE 2. Ribbon representation of ligand-free PikC. A,
open, and B, closed conformations of ligand-free PikC (2BVJ). C,
overlay of both conformations, open (cyan) and closed (gray),
demonstrating that in the open form, the F and G helix are bent
away from the heme to enable substrate access to the active
site. Molecules in C are rotated  90° toward the viewer
along a horizontal axis in the plane of drawing when compared
with A and B. The F helix is not seen in this orientation.
Closed conformation is related within r.m.s. deviations of 0.58
Å for C 90° toward the viewer
along a horizontal axis in the plane of drawing when compared
with A and B. The F helix is not seen in this orientation.
Closed conformation is related within r.m.s. deviations of 0.58
Å for C  atoms to catalytically
relevant YC-17- and narbomycin-bound forms. The heme co-factor
is shown in red. atoms to catalytically
relevant YC-17- and narbomycin-bound forms. The heme co-factor
is shown in red.
|
 |
Figure 6.
FIGURE 6. Functional activity of PikC mutants. High
pressure liquid chromatography analyses of PikC-catalyzed
reactions using YC-17 (A series) and narbomycin (B series) as
substrate are shown. A1/B1, negative control in the absence of
PikC. A2/B2, PikC wild type (PikC-wt). Mutants are used as
indicated in the figure. Compound identities are as follows: 1,
YC-17; 2, neomethymycin; 3, methymycin; 4, narbomycin; 5,
pikromycin. Conversion of narbomycin at low efficiency is
probably due to use of exogenous redox partners (e.g. spinach
ferredoxin reductase) in P450 reconstitution assays.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
26289-26297)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Ma,
S.G.Bell,
W.Yang,
Y.Hao,
N.H.Rees,
M.Bartlam,
W.Zhou,
L.L.Wong,
and
Z.Rao
(2011).
Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
|
| |
Chembiochem,
12,
88-99.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.R.Markwick,
L.C.Pierce,
D.B.Goodin,
and
J.A.McCammon
(2011).
Adaptive Accelerated Molecular Dynamics (Ad-AMD) Revealing the Molecular Plasticity of P450cam.
|
| |
J Phys Chem Lett,
2,
158-164.
|
 |
|
|
|
|
 |
Y.T.Lee,
E.C.Glazer,
R.F.Wilson,
C.D.Stout,
and
D.B.Goodin
(2011).
Three clusters of conformational States in p450cam reveal a multistep pathway for closing of the substrate access channel .
|
| |
Biochemistry,
50,
693-703.
|
 |
|
|
|
|
 |
C.Olano,
C.Méndez,
and
J.A.Salas
(2010).
Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis.
|
| |
Nat Prod Rep,
27,
571-616.
|
 |
|
|
|
|
 |
S.R.Park,
A.R.Han,
Y.H.Ban,
Y.J.Yoo,
E.J.Kim,
and
Y.J.Yoon
(2010).
Genetic engineering of macrolide biosynthesis: past advances, current state, and future prospects.
|
| |
Appl Microbiol Biotechnol,
85,
1227-1239.
|
 |
|
|
|
|
 |
T.C.Pochapsky,
S.Kazanis,
and
M.Dang
(2010).
Conformational plasticity and structure/function relationships in cytochromes P450.
|
| |
Antioxid Redox Signal,
13,
1273-1296.
|
 |
|
|
|
|
 |
Y.T.Lee,
R.F.Wilson,
I.Rupniewski,
and
D.B.Goodin
(2010).
P450cam visits an open conformation in the absence of substrate.
|
| |
Biochemistry,
49,
3412-3419.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Hong,
S.G.Bell,
W.Yang,
H.Wang,
Y.Hao,
X.Li,
W.Zhou,
M.Bartlam,
and
L.L.Wong
(2009).
Purification, crystallization and preliminary X-ray analysis of cytochrome P450 219A1 from Novosphingobium aromaticivorans DSM 12444.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
364-367.
|
 |
|
|
|
|
 |
C.Savino,
L.C.Montemiglio,
G.Sciara,
A.E.Miele,
S.G.Kendrew,
P.Jemth,
S.Gianni,
and
B.Vallone
(2009).
Investigating the structural plasticity of a cytochrome P450: three-dimensional structures of P450 EryK and binding to its physiological substrate.
|
| |
J Biol Chem,
284,
29170-29179.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.G.Denisov,
D.J.Frank,
and
S.G.Sligar
(2009).
Cooperative properties of cytochromes P450.
|
| |
Pharmacol Ther,
124,
151-167.
|
 |
|
|
|
|
 |
J.G.McCoy,
H.D.Johnson,
S.Singh,
C.A.Bingman,
I.K.Lei,
J.S.Thorson,
and
G.N.Phillips
(2009).
Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
|
| |
Proteins,
74,
50-60.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.H.Xu,
S.Fushinobu,
H.Ikeda,
T.Wakagi,
and
H.Shoun
(2009).
Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state.
|
| |
J Bacteriol,
191,
1211-1219.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Li,
H.Ouellet,
D.H.Sherman,
and
L.M.Podust
(2009).
Analysis of transient and catalytic desosamine-binding pockets in cytochrome P-450 PikC from Streptomyces venezuelae.
|
| |
J Biol Chem,
284,
5723-5730.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Li,
M.R.Chaulagain,
A.R.Knauff,
L.M.Podust,
J.Montgomery,
and
D.H.Sherman
(2009).
Selective oxidation of carbolide C-H bonds by an engineered macrolide P450 mono-oxygenase.
|
| |
Proc Natl Acad Sci U S A,
106,
18463-18468.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Yasutake,
Y.Fujii,
W.K.Cheon,
A.Arisawa,
and
T.Tamura
(2009).
Crystallization and preliminary X-ray diffraction studies of vitamin D3 hydroxylase, a novel cytochrome P450 isolated from Pseudonocardia autotrophica.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
372-375.
|
 |
|
|
|
|
 |
H.Ouellet,
L.M.Podust,
and
P.R.de Montellano
(2008).
Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs.
|
| |
J Biol Chem,
283,
5069-5080.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Cryle,
and
I.Schlichting
(2008).
Structural insights from a P450 Carrier Protein complex reveal how specificity is achieved in the P450(BioI) ACP complex.
|
| |
Proc Natl Acad Sci U S A,
105,
15696-15701.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Shrestha,
T.J.Oh,
K.Liou,
and
J.K.Sohng
(2008).
Cytochrome P450 (CYP105F2) from Streptomyces peucetius and its activity with oleandomycin.
|
| |
Appl Microbiol Biotechnol,
79,
555-562.
|
 |
|
|
|
|
 |
W.S.Jung,
S.J.Jeong,
S.R.Park,
C.Y.Choi,
B.C.Park,
J.W.Park,
and
Y.J.Yoon
(2008).
Enhanced heterologous production of desosaminyl macrolides and their hydroxylated derivatives by overexpression of the pikD regulatory gene in Streptomyces venezuelae.
|
| |
Appl Environ Microbiol,
74,
1972-1979.
|
 |
|
|
|
|
 |
Y.Anzai,
S.Li,
M.R.Chaulagain,
K.Kinoshita,
F.Kato,
J.Montgomery,
and
D.H.Sherman
(2008).
Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics.
|
| |
Chem Biol,
15,
950-959.
|
 |
|
|
|
|
 |
A.W.Munro,
H.M.Girvan,
and
K.J.McLean
(2007).
Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily.
|
| |
Nat Prod Rep,
24,
585-609.
|
 |
|
|
|
|
 |
S.Li,
L.M.Podust,
and
D.H.Sherman
(2007).
Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain.
|
| |
J Am Chem Soc,
129,
12940-12941.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

