 |
PDBsum entry 1vao

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.38
- vanillyl-alcohol oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxy-3-methoxy-benzenemethanol + O2 = vanillin + H2O2
|
 |
 |
 |
 |
 |
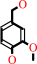 4-hydroxy-3-methoxy-benzenemethanol
4-hydroxy-3-methoxy-benzenemethanol
|
+
|
 O2
O2
|
=
|
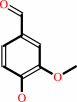 vanillin
vanillin
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Structure
5:907-920
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures and inhibitor binding in the octameric flavoenzyme vanillyl-alcohol oxidase: the shape of the active-site cavity controls substrate specificity.
|
|
A.Mattevi,
M.W.Fraaije,
A.Mozzarelli,
L.Olivi,
A.Coda,
W.J.van Berkel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Lignin degradation leads to the formation of a broad spectrum of
aromatic molecules that can be used by various fungal micro-organisms as their
sole source of carbon. When grown on phenolic compounds, Penicillium
simplicissimum induces the strong impression of a flavin-containing
vanillyl-alcohol oxidase (VAO). The enzyme catalyses the oxidation of a vast
array of substrates, ranging from aromatic amines to 4-alkyphenols. VAO is a
member of a novel class of widely distributed oxidoreductases, which use flavin
adenine dinucleotide (FAD) as a cofactor covalently bound to the protein. We
have carried out the determination of the structure of VAO in order to shed
light on the most interesting features of these novel oxidoreductases, such as
the functional significance of covalent flavinylation and the mechanism of
catalysis. RESULTS: The crystal structure of VAO has been determined in the
native state and in complexes with four inhibitors. The enzyme is an octamer
with 42 symmetry; the inhibitors bind in a hydrophobic, elongated cavity on the
si side of the flavin molecule. Three residues, Tyr108, Tyr503 and Arg504 form
an anion-binding subsite, which stabilises the phenolate form of the substrate.
The structure of VAO complexed with the inhibitor 4-(1-heptenyl)phenol shows
that the catalytic cavity is completely filled by the inhibitor, explaining why
alkylphenols bearing aliphatic substituents longer than seven carbon atoms do
not bind to the enzyme. CONCLUSIONS: The shape of the active-site cavity
controls substrate specificity by providing a 'size exclusion mechanism'. Inside
the cavity, the substrate aromatic ring is positioned at an angle of 18 degrees
to the flavin ring. This arrangement is ideally suited for a hydride transfer
reaction, which is further facilitated by substrate deprotonation. Burying the
substrate beneath the protein surface is a recurrent strategy, common to many
flavoenzymes that effect substrate oxidation or reduction via hydride transfer.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Janner
(2010).
Form, symmetry and packing of biomacromolecules. I. Concepts and tutorial examples.
|
| |
Acta Crystallogr A,
66,
301-311.
|
 |
|
|
|
|
 |
L.M.Blank,
B.E.Ebert,
K.Buehler,
and
B.Bühler
(2010).
Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis.
|
| |
Antioxid Redox Signal,
13,
349-394.
|
 |
|
|
|
|
 |
F.Forneris,
R.Orru,
D.Bonivento,
L.R.Chiarelli,
and
A.Mattevi
(2009).
ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding.
|
| |
FEBS J,
276,
2833-2840.
|
 |
|
|
|
|
 |
I.S.Fernández,
F.J.Ruíz-Dueñas,
E.Santillana,
P.Ferreira,
M.J.Martínez,
A.T.Martínez,
and
A.Romero
(2009).
Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1196-1205.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Siltberg-Liberles,
and
A.Martinez
(2009).
Searching distant homologs of the regulatory ACT domain in phenylalanine hydroxylase.
|
| |
Amino Acids,
36,
235-249.
|
 |
|
|
|
|
 |
N.G.Leferink,
M.W.Fraaije,
H.J.Joosten,
P.J.Schaap,
A.Mattevi,
and
W.J.van Berkel
(2009).
Identification of a gatekeeper residue that prevents dehydrogenases from acting as oxidases.
|
| |
J Biol Chem,
284,
4392-4397.
|
 |
|
|
|
|
 |
G.Zhao,
R.C.Bruckner,
and
M.S.Jorns
(2008).
Identification of the oxygen activation site in monomeric sarcosine oxidase: role of Lys265 in catalysis.
|
| |
Biochemistry,
47,
9124-9135.
|
 |
|
|
|
|
 |
J.Jin,
H.Mazon,
R.H.van den Heuvel,
A.J.Heck,
D.B.Janssen,
and
M.W.Fraaije
(2008).
Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process.
|
| |
FEBS J,
275,
5191-5200.
|
 |
|
|
|
|
 |
J.Jin,
H.Mazon,
R.H.van den Heuvel,
D.B.Janssen,
and
M.W.Fraaije
(2007).
Discovery of a eugenol oxidase from Rhodococcus sp. strain RHA1.
|
| |
FEBS J,
274,
2311-2321.
|
 |
|
|
|
|
 |
A.Mattevi
(2006).
To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes.
|
| |
Trends Biochem Sci,
31,
276-283.
|
 |
|
|
|
|
 |
P.Ferreira,
F.J.Ruiz-Dueñas,
M.J.Martínez,
W.J.van Berkel,
and
A.T.Martínez
(2006).
Site-directed mutagenesis of selected residues at the active site of aryl-alcohol oxidase, an H2O2-producing ligninolytic enzyme.
|
| |
FEBS J,
273,
4878-4888.
|
 |
|
|
|
|
 |
A.Janner
(2005).
Strongly correlated structure of axial-symmetric proteins. I. Orthorhombic, tetragonal, trigonal and hexagonal symmetries.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
247-255.
|
 |
|
|
|
|
 |
A.J.Heck,
and
R.H.Van Den Heuvel
(2004).
Investigation of intact protein complexes by mass spectrometry.
|
| |
Mass Spectrom Rev,
23,
368-389.
|
 |
|
|
|
|
 |
R.H.van den Heuvel,
W.A.van den Berg,
S.Rovida,
and
W.J.van Berkel
(2004).
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin.
|
| |
J Biol Chem,
279,
33492-33500.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Tahallah,
R.H.Van Den Heuvel,
W.A.Van Den Berg,
C.S.Maier,
W.J.Van Berkel,
and
A.J.Heck
(2002).
Cofactor-dependent assembly of the flavoenzyme vanillyl-alcohol oxidase.
|
| |
J Biol Chem,
277,
36425-36432.
|
 |
|
|
|
|
 |
C.Breithaupt,
J.Strassner,
U.Breitinger,
R.Huber,
P.Macheroux,
A.Schaller,
and
T.Clausen
(2001).
X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
|
| |
Structure,
9,
419-429.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.E.Edmondson,
and
P.Newton-Vinson
(2001).
The covalent FAD of monoamine oxidase: structural and functional role and mechanism of the flavinylation reaction.
|
| |
Antioxid Redox Signal,
3,
789-806.
|
 |
|
|
|
|
 |
I.Efimov,
C.N.Cronin,
and
W.S.McIntire
(2001).
Effects of noncovalent and covalent FAD binding on the redox and catalytic properties of p-cresol methylhydroxylase.
|
| |
Biochemistry,
40,
2155-2166.
|
 |
|
|
|
|
 |
M.D.Krasowski,
K.Nishikawa,
N.Nikolaeva,
A.Lin,
and
N.L.Harrison
(2001).
Methionine 286 in transmembrane domain 3 of the GABAA receptor beta subunit controls a binding cavity for propofol and other alkylphenol general anesthetics.
|
| |
Neuropharmacology,
41,
952-964.
|
 |
|
|
|
|
 |
N.Tahallah,
M.Pinkse,
C.S.Maier,
and
A.J.Heck
(2001).
The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument.
|
| |
Rapid Commun Mass Spectrom,
15,
596-601.
|
 |
|
|
|
|
 |
S.Peri,
N.Ibarrola,
B.Blagoev,
M.Mann,
and
A.Pandey
(2001).
Common pitfalls in bioinformatics-based analyses: look before you leap.
|
| |
Trends Genet,
17,
541-545.
|
 |
|
|
|
|
 |
T.E.Benson,
M.S.Harris,
G.H.Choi,
J.I.Cialdella,
J.T.Herberg,
J.P.Martin,
and
E.T.Baldwin
(2001).
A structural variation for MurB: X-ray crystal structure of Staphylococcus aureus UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
|
| |
Biochemistry,
40,
2340-2350.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.C.de Vet,
Y.H.Hilkes,
M.W.Fraaije,
and
H.van den Bosch
(2000).
Alkyl-dihydroxyacetonephosphate synthase. Presence and role of flavin adenine dinucleotide.
|
| |
J Biol Chem,
275,
6276-6283.
|
 |
|
|
|
|
 |
E.Varela,
M.Jesús Martínez,
and
A.T.Martínez
(2000).
Aryl-alcohol oxidase protein sequence: a comparison with glucose oxidase and other FAD oxidoreductases.
|
| |
Biochim Biophys Acta,
1481,
202-208.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Gadda,
and
P.F.Fitzpatrick
(2000).
Mechanism of nitroalkane oxidase: 2. pH and kinetic isotope effects.
|
| |
Biochemistry,
39,
1406-1410.
|
 |
|
|
|
|
 |
M.A.Wagner,
P.Trickey,
Z.W.Chen,
F.S.Mathews,
and
M.S.Jorns
(2000).
Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
|
| |
Biochemistry,
39,
8813-8824.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.J.Walton,
A.Narbad,
C.Faulds,
and
G.Williamson
(2000).
Novel approaches to the biosynthesis of vanillin.
|
| |
Curr Opin Biotechnol,
11,
490-496.
|
 |
|
|
|
|
 |
O.Dym,
E.A.Pratt,
C.Ho,
and
D.Eisenberg
(2000).
The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
|
| |
Proc Natl Acad Sci U S A,
97,
9413-9418.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.H.van Den Heuvel,
M.W.Fraaije,
M.Ferrer,
A.Mattevi,
and
W.J.van Berkel
(2000).
Inversion of stereospecificity of vanillyl-alcohol oxidase.
|
| |
Proc Natl Acad Sci U S A,
97,
9455-9460.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.H.van den Heuvel,
M.W.Fraaije,
A.Mattevi,
and
W.J.van Berkel
(2000).
Asp-170 is crucial for the redox properties of vanillyl-alcohol oxidase.
|
| |
J Biol Chem,
275,
14799-14808.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.J.van Berkel,
R.H.van den Heuvel,
C.Versluis,
and
A.J.Heck
(2000).
Detection of intact megaDalton protein assemblies of vanillyl-alcohol oxidase by mass spectrometry.
|
| |
Protein Sci,
9,
435-439.
|
 |
|
|
|
|
 |
A.Mattevi,
G.Tedeschi,
L.Bacchella,
A.Coda,
A.Negri,
and
S.Ronchi
(1999).
Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family.
|
| |
Structure,
7,
745-756.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Oubrie,
H.J.Rozeboom,
K.H.Kalk,
A.J.Olsthoorn,
J.A.Duine,
and
B.W.Dijkstra
(1999).
Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
|
| |
EMBO J,
18,
5187-5194.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Binda,
A.Coda,
R.Angelini,
R.Federico,
P.Ascenzi,
and
A.Mattevi
(1999).
A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
|
| |
Structure,
7,
265-276.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Dobbek,
L.Gremer,
O.Meyer,
and
R.Huber
(1999).
Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine.
|
| |
Proc Natl Acad Sci U S A,
96,
8884-8889.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Bacchella,
C.Lina,
F.Todone,
A.Negri,
G.Tedeschi,
S.Ronchi,
and
A.Mattevi
(1999).
Crystallization of L-aspartate oxidase, the first enzyme in the bacterial de novo biosynthesis of NAD.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
549-551.
|
 |
|
|
|
|
 |
M.W.Fraaije,
R.H.van den Heuvel,
W.J.van Berkel,
and
A.Mattevi
(1999).
Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase.
|
| |
J Biol Chem,
274,
35514-35520.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Trickey,
M.A.Wagner,
M.S.Jorns,
and
F.S.Mathews
(1999).
Monomeric sarcosine oxidase: structure of a covalently flavinylated amine oxidizing enzyme.
|
| |
Structure,
7,
331-345.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Mattevi
(1998).
The PHBH fold: not only flavoenzymes.
|
| |
Biophys Chem,
70,
217-222.
|
 |
|
|
|
|
 |
C.Binda,
A.Coda,
R.Angelini,
R.Federico,
P.Ascenzi,
and
A.Mattevi
(1998).
Crystallization and preliminary X-ray analysis of polyamine oxidase from Zea mays L.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
1429-1431.
|
 |
|
|
|
|
 |
F.P.Drijfhout,
M.W.Fraaije,
H.Jongejan,
van Berkel WJ,
and
M.C.Franssen
(1998).
Enantioselective hydroxylation of 4-alkylphenols by vanillyl alcohol oxidase
|
| |
Biotechnol Bioeng,
59,
171-177.
|
 |
|
|
|
|
 |
J.A.Benen,
P.Sánchez-Torres,
M.J.Wagemaker,
M.W.Fraaije,
W.J.van Berkel,
and
J.Visser
(1998).
Molecular cloning, sequencing, and heterologous expression of the vaoA gene from Penicillium simplicissimum CBS 170.90 encoding vanillyl-alcohol oxidase.
|
| |
J Biol Chem,
273,
7865-7872.
|
 |
|
|
|
|
 |
L.Holm
(1998).
Unification of protein families.
|
| |
Curr Opin Struct Biol,
8,
372-379.
|
 |
|
|
|
|
 |
M.W.Fraaije,
W.J.Van Berkel,
J.A.Benen,
J.Visser,
and
A.Mattevi
(1998).
A novel oxidoreductase family sharing a conserved FAD-binding domain.
|
| |
Trends Biochem Sci,
23,
206-207.
|
 |
|
|
|
|
 |
R.H.van den Heuvel,
M.W.Fraaije,
C.Laane,
and
W.J.van Berkel
(1998).
Regio- and stereospecific conversion of 4-alkylphenols by the covalent flavoprotein vanillyl-alcohol oxidase.
|
| |
J Bacteriol,
180,
5646-5651.
|
 |
|
|
|
|
 |
A.Mattevi,
M.A.Vanoni,
and
B.Curti
(1997).
Structure of D-amino acid oxidase: new insights from an old enzyme.
|
| |
Curr Opin Struct Biol,
7,
804-810.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

