 |
PDBsum entry 1f0x

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1f0x
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.5.12
- D-lactate dehydrogenase (quinone).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-lactate + a quinone = a quinol + pyruvate
|
 |
 |
 |
 |
 |
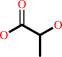 (R)-lactate
(R)-lactate
|
+
|
 quinone
quinone
|
=
|
quinol
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
97:9413-9418
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
|
|
O.Dym,
E.A.Pratt,
C.Ho,
D.Eisenberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
d-Lactate dehydrogenase (d-LDH) of Escherichia coli is a peripheral membrane
respiratory enzyme involved in electron transfer, located on the cytoplasmic
side of the inner membrane. d-LDH catalyzes the oxidation of d-lactate to
pyruvate, which is coupled to transmembrane transport of amino acids and sugars.
Here we describe the crystal structure at 1.9 A resolution of the three domains
of d-LDH: the flavin adenine dinucleotide (FAD)-binding domain, the cap domain,
and the membrane-binding domain. The FAD-binding domain contains the site of
d-lactate reduction by a noncovalently bound FAD cofactor and has an overall
fold similar to other members of a recently discovered FAD-containing family of
proteins. This structural similarity extends to the cap domain as well. The most
prominent difference between d-LDH and the other members of the FAD-containing
family is the membrane-binding domain, which is either absent in some of these
proteins or differs significantly. The d-LDH membrane-binding domain presents an
electropositive surface with six Arg and five Lys residues, which presumably
interacts with the negatively charged phospholipid head groups of the membrane.
Thus, d-LDH appears to bind the membrane through electrostatic rather than
hydrophobic forces.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Schematic diagram of D-LDH interactions with the
FAD cofactor was created by using the program LIGPLOT (42). All
van der Waals interactions and hydrogen bond contacts to the FAD
cofactor (Middle) are contributed solely from residues of the
FAD-binding domain. The residues that form hydrogen-bonds to the
FAD are shown in ball-and-stick representation. Hydrogen bonds
are presented as dashed lines and the interatomic distances are
shown in angstroms. The residues that form van der Waals
contacts with the FAD are depicted as labeled arcs with radial
spokes that point toward the ligand atoms with which they
interact.
|
 |
Figure 4.
Fig. 4. Cartoon of D-LDH associating with the membrane.
D-LDH is anchored to the membrane by electrostatic interactions
between basic residues (blue balls) from the observed
membrane-binding domain (blue) and possibly from the modeled
missing segment (dashed yellow) comprising nine basic residues
(yellow balls) and the negatively charged phospholipid head
groups (red balls) of the membrane. The cap domain (pink) and
FAD-binding domain (cyan) are also shown. The stick drawing of
the FAD cofactor is depicted with gray balls for atoms in the
adenine and sugar rings, with red balls for phosphate and oxygen
atoms, and with black balls for atoms in the isoalloxazine ring.
In this model, the substrate D-lactate can approach the active
site (as shown by the black arrow) near the isoalloxazine ring
(visible between pink strands) from behind the cap domain.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.T.Thomas,
M.Shepherd,
R.K.Poole,
A.H.van Vliet,
D.J.Kelly,
and
B.M.Pearson
(2011).
Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate.
|
| |
Environ Microbiol,
13,
48-61.
|
 |
|
|
|
|
 |
M.Wang,
Y.Y.Jiang,
K.M.Kim,
G.Qu,
H.F.Ji,
J.E.Mittenthal,
H.Y.Zhang,
and
G.Caetano-Anollés
(2011).
A universal molecular clock of protein folds and its power in tracing the early history of aerobic metabolism and planet oxygenation.
|
| |
Mol Biol Evol,
28,
567-582.
|
 |
|
|
|
|
 |
F.R.Laham,
A.A.Trott,
B.L.Bennett,
C.A.Kozinetz,
A.M.Jewell,
R.P.Garofalo,
and
P.A.Piedra
(2010).
LDH concentration in nasal-wash fluid as a biochemical predictor of bronchiolitis severity.
|
| |
Pediatrics,
125,
e225-e233.
|
 |
|
|
|
|
 |
O.Kato,
J.W.Youn,
K.C.Stansen,
D.Matsui,
T.Oikawa,
and
V.F.Wendisch
(2010).
Quinone-dependent D-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on D-lactate.
|
| |
BMC Microbiol,
10,
321.
|
 |
|
|
|
|
 |
J.M.Liefhebber,
B.W.Brandt,
R.Broer,
W.J.Spaan,
and
H.C.van Leeuwen
(2009).
Hepatitis C virus NS4B carboxy terminal domain is a membrane binding domain.
|
| |
Virol J,
6,
62.
|
 |
|
|
|
|
 |
M.E.Cristescu,
and
E.E.Egbosimba
(2009).
Evolutionary history of D-lactate dehydrogenases: a phylogenomic perspective on functional diversity in the FAD binding oxidoreductase/transferase type 4 family.
|
| |
J Mol Evol,
69,
276-287.
|
 |
|
|
|
|
 |
A.Razeto,
F.Mattiroli,
E.Carpanelli,
A.Aliverti,
V.Pandini,
A.Coda,
and
A.Mattevi
(2007).
The crucial step in ether phospholipid biosynthesis: structural basis of a noncanonical reaction associated with a peroxisomal disorder.
|
| |
Structure,
15,
683-692.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Ma,
C.Gao,
J.Qiu,
J.Hao,
W.Liu,
A.Wang,
Y.Zhang,
M.Wang,
and
P.Xu
(2007).
Membrane-bound L- and D-lactate dehydrogenase activities of a newly isolated Pseudomonas stutzeri strain.
|
| |
Appl Microbiol Biotechnol,
77,
91-98.
|
 |
|
|
|
|
 |
J.B.McKinlay,
C.Vieille,
and
J.G.Zeikus
(2007).
Prospects for a bio-based succinate industry.
|
| |
Appl Microbiol Biotechnol,
76,
727-740.
|
 |
|
|
|
|
 |
L.Durant,
A.Metais,
C.Soulama-Mouze,
J.M.Genevard,
X.Nassif,
and
S.Escaich
(2007).
Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli.
|
| |
Infect Immun,
75,
1916-1925.
|
 |
|
|
|
|
 |
R.J.Siezen,
B.Renckens,
I.van Swam,
S.Peters,
R.van Kranenburg,
M.Kleerebezem,
and
W.M.de Vos
(2005).
Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment.
|
| |
Appl Environ Microbiol,
71,
8371-8382.
|
 |
|
|
|
|
 |
P.Goffin,
F.Lorquet,
M.Kleerebezem,
and
P.Hols
(2004).
Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase.
|
| |
J Bacteriol,
186,
6661-6666.
|
 |
|
|
|
|
 |
J.Sivaraman,
V.Sauvé,
R.Larocque,
E.A.Stura,
J.D.Schrag,
M.Cygler,
and
A.Matte
(2002).
Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
|
| |
Nat Struct Biol,
9,
353-358.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.J.White,
R.Merod,
B.Thomasson,
and
P.L.Hartzell
(2001).
GidA is an FAD-binding protein involved in development of Myxococcus xanthus.
|
| |
Mol Microbiol,
42,
503-517.
|
 |
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

