 |
PDBsum entry 1v51

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.81
- N-acyl-D-amino-acid deacylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acyl-D-amino acid + H2O = a D-alpha-amino acid + a carboxylate
|
 |
 |
 |
 |
 |
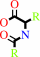 N-acyl-D-amino acid
N-acyl-D-amino acid
|
+
|
H2O
|
=
|
D-alpha-amino acid
Bound ligand (Het Group name = )
matches with 60.00% similarity
|
+
|
 carboxylate
carboxylate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:13962-13967
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
|
|
W.L.Lai,
L.Y.Chou,
C.Y.Ting,
R.Kirby,
Y.C.Tsai,
A.H.Wang,
S.H.Liaw.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Our structural comparison of the TIM barrel metal-dependent hydrolase(-like)
superfamily suggests a classification of their divergent active sites into four
types: alphabeta-binuclear, alpha-mononuclear, beta-mononuclear, and
metal-independent subsets. The d-aminoacylase from Alcaligenes faecalis DA1
belongs to the beta-mononuclear subset due to the fact that the catalytically
essential Zn(2+) is tightly bound at the beta site with coordination by Cys(96),
His(220), and His(250), even though it possesses a binuclear active site with a
weak alpha binding site. Additional Zn(2+), Cd(2+), and Cu(2+), but not Ni(2+),
Co(2+), Mg(2+), Mn(2+), and Ca(2+), can inhibit enzyme activity. Crystal
structures of these metal derivatives show that Zn(2+) and Cd(2+) bind at the
alpha(1) subsite ligated by His(67), His(69), and Asp(366), while Cu(2+) at the
alpha(2) subsite is chelated by His(67), His(69) and Cys(96). Unexpectedly, the
crystal structure of the inactive H220A mutant displays that the endogenous
Zn(2+) shifts to the alpha(3) subsite coordinated by His(67), His(69), Cys(96),
and Asp(366), revealing that elimination of the beta site changes the
coordination geometry of the alpha ion with an enhanced affinity. Kinetic
studies of the metal ligand mutants such as C96D indicate the uniqueness of the
unusual bridging cysteine and its involvement in catalysis. Therefore, the two
metal-binding sites in the d-aminoacylase are interactive with partially mutual
exclusion, thus resulting in widely different affinities for the
activation/attenuation mechanism, in which the enzyme is activated by the metal
ion at the beta site, but inhibited by the subsequent binding of the second ion
at the alpha site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIG. 3. The metal centers. A, the F[o] - F[c] electron
density maps of the native enzyme in complex with 100 mM ZnCl[2]
contoured at 15  level and shown in
magenta, with 50 mM CdCl[2] contoured at 15 level and shown in
magenta, with 50 mM CdCl[2] contoured at 15  level and shown in
cyan, and with 100 mM CuCl[2] contoured at 18 level and shown in
cyan, and with 100 mM CuCl[2] contoured at 18  level
and shown in green. The metal ligands are shown as a
ball-and-stick representation, with the Zn2+ and Cu2+ ions as
magenta and green spheres, respectively. Zn2+ and Cd^2+ bind at
the level
and shown in green. The metal ligands are shown as a
ball-and-stick representation, with the Zn2+ and Cu2+ ions as
magenta and green spheres, respectively. Zn2+ and Cd^2+ bind at
the  subsite, where Cu2+
binds at the subsite, where Cu2+
binds at the  [2] subsite. B, the
2F[o] - F[c] electron density maps of the H220A mutant contoured
at 2.5 [2] subsite. B, the
2F[o] - F[c] electron density maps of the H220A mutant contoured
at 2.5  level and shown in
cyan, and the difference map for the zinc ion contoured at 15 level and shown in
cyan, and the difference map for the zinc ion contoured at 15
 level and shown in
magenta. The endogenous zinc ion binds at the level and shown in
magenta. The endogenous zinc ion binds at the  [3]
subsite instead of the [3]
subsite instead of the  site in this mutant. C,
the 2F[o] - F[c] electron density map of the D366A mutant
contoured at 2.5 site in this mutant. C,
the 2F[o] - F[c] electron density map of the D366A mutant
contoured at 2.5  level and shown in
cyan, and the difference map for the zinc ion in complex with
100 mM ZnCl[2] contoured at 15 level and shown in
cyan, and the difference map for the zinc ion in complex with
100 mM ZnCl[2] contoured at 15  level and shown in
magenta. The additional zinc ion binds at the level and shown in
magenta. The additional zinc ion binds at the  [4]
subsite. D, superposition of the native enzyme with 100 mM
ZnCl[2] in blue, the native enzyme with 100 CuCl[2] in green,
the H220A mutant in yellow, and the D366A mutant with 100 mM
ZnCl[2] in red. The different metal coordination is carried out
by small shifts in the side chains of ligands and small
movements of the metal ions. [4]
subsite. D, superposition of the native enzyme with 100 mM
ZnCl[2] in blue, the native enzyme with 100 CuCl[2] in green,
the H220A mutant in yellow, and the D366A mutant with 100 mM
ZnCl[2] in red. The different metal coordination is carried out
by small shifts in the side chains of ligands and small
movements of the metal ions.
|
 |
Figure 4.
FIG. 4. The proposed mechanisms for catalysis (A) and metal
attenuation (B). The numbers shown indicate the interatomic
distances in angstroms. Asp366 maybe with assistance from His67
and His69, is responsible for the proton transfer from the water
molecule to the amide nitrogen (3). The presence of the
inhibitory metal ion at the  [1] site might lower
the pK[a] values of its ligand residues, His67, His69, and
Asp366, and/or hold the active site water to perturb the proton
shuttle and intermediate stabilization. [1] site might lower
the pK[a] values of its ligand residues, His67, His69, and
Asp366, and/or hold the active site water to perturb the proton
shuttle and intermediate stabilization.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
13962-13967)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Latorre,
F.Olivares,
A.Reyes-Jara,
G.López,
and
M.González
(2011).
CutC is induced late during copper exposure and can modify intracellular copper content in Enterococcus faecalis.
|
| |
Biochem Biophys Res Commun,
406,
633-637.
|
 |
|
|
|
|
 |
J.A.Cummings,
A.A.Fedorov,
C.Xu,
S.Brown,
E.Fedorov,
P.C.Babbitt,
S.C.Almo,
and
F.M.Raushel
(2009).
Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
|
| |
Biochemistry,
48,
6469-6481.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Palaniappan,
V.Dhote,
S.Ayers,
A.L.Starosta,
D.N.Wilson,
and
K.A.Reynolds
(2009).
Biosynthesis of the aminocyclitol subunit of hygromycin A in Streptomyces hygroscopicus NRRL 2388.
|
| |
Chem Biol,
16,
1180-1189.
|
 |
|
|
|
|
 |
S.Wydau,
G.van der Rest,
C.Aubard,
P.Plateau,
and
S.Blanquet
(2009).
Widespread distribution of cell defense against D-aminoacyl-tRNAs.
|
| |
J Biol Chem,
284,
14096-14104.
|
 |
|
|
|
|
 |
J.Kim,
P.C.Tsai,
S.L.Chen,
F.Himo,
S.C.Almo,
and
F.M.Raushel
(2008).
Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
|
| |
Biochemistry,
47,
9497-9504.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Tanimoto,
N.Higashi,
M.Nishioka,
K.Ishikawa,
and
M.Taya
(2008).
Characterization of thermostable aminoacylase from hyperthermophilic archaeon Pyrococcus horikoshii.
|
| |
FEBS J,
275,
1140-1149.
|
 |
|
|
|
|
 |
M.Babor,
S.Gerzon,
B.Raveh,
V.Sobolev,
and
M.Edelman
(2008).
Prediction of transition metal-binding sites from apo protein structures.
|
| |
Proteins,
70,
208-217.
|
 |
|
|
|
|
 |
R.E.Mirams,
S.J.Smith,
K.S.Hadler,
D.L.Ollis,
G.Schenk,
and
L.R.Gahan
(2008).
Cadmium(II) complexes of the glycerophosphodiester-degrading enzyme GpdQ and a biomimetic N,O ligand.
|
| |
J Biol Inorg Chem,
13,
1065-1072.
|
 |
|
|
|
|
 |
T.Dudev,
and
C.Lim
(2008).
Metal binding affinity and selectivity in metalloproteins: insights from computational studies.
|
| |
Annu Rev Biophys,
37,
97.
|
 |
|
|
|
|
 |
J.M.Xu,
F.Zhang,
B.K.Liu,
Q.Wu,
and
X.F.Lin
(2007).
Promiscuous zinc-dependent acylase-mediated carbon-carbon bond formation in organic media.
|
| |
Chem Commun (Camb),
(),
2078-2080.
|
 |
|
|
|
|
 |
K.Yoshimune,
A.Hirayama,
and
M.Moriguchi
(2005).
A metal ion as a cofactor attenuates substrate inhibition in the enzymatic production of a high concentration of D-glutamate using N-acyl-D-glutamate amidohydrolase.
|
| |
Biotechnol Lett,
27,
1325-1328.
|
 |
|
|
|
|
 |
S.Deejing,
K.Yoshimune,
S.Lumyong,
and
M.Moriguchi
(2005).
Purification and characterization of hyperthermotolerant leucine aminopeptidase from Geobacillus thermoleovorans 47b.
|
| |
J Ind Microbiol Biotechnol,
32,
269-276.
|
 |
|
|
|
|
 |
Y.J.Chang,
C.H.Huang,
C.Y.Hu,
and
S.H.Liaw
(2004).
Crystallization and preliminary crystallographic analysis of Bacillus subtilis guanine deaminase.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1152-1154.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

