 |
PDBsum entry 1pyd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase(carbon-carbon)
|
PDB id
|
|

|

|
1pyd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.1.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.4.1.1.43
- phenylpyruvate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-phenylpyruvate + H+ = 2-phenylacetaldehyde + CO2
|
 |
 |
 |
 |
 |
3-phenylpyruvate
|
+
|
H(+)
|
=
|
2-phenylacetaldehyde
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.4.1.1.72
- branched-chain-2-oxoacid decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-3-methyl-2-oxopentanoate + H+ = 2-methylbutanal + CO2
|
 |
 |
 |
 |
 |
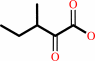 (S)-3-methyl-2-oxopentanoate
(S)-3-methyl-2-oxopentanoate
|
+
|
H(+)
|
=
|
 2-methylbutanal
2-methylbutanal
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.4.1.1.74
- indolepyruvate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
indole-3-pyruvate + H+ = indole-3-acetaldehyde + CO2
|
 |
 |
 |
 |
 |
 indole-3-pyruvate
indole-3-pyruvate
|
+
|
H(+)
|
=
|
 indole-3-acetaldehyde
indole-3-acetaldehyde
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+); Thiamine diphosphate
|
 |
 |
 |
 |
 |
Mg(2+)
|
Thiamine diphosphate
Bound ligand (Het Group name =
TPP)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:6165-6170
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Catalytic centers in the thiamin diphosphate dependent enzyme pyruvate decarboxylase at 2.4-A resolution.
|
|
F.Dyda,
W.Furey,
S.Swaminathan,
M.Sax,
B.Farrenkopf,
F.Jordan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of brewers' yeast pyruvate decarboxylase, a thiamin
diphosphate dependent alpha-keto acid decarboxylase, has been determined to
2.4-A resolution. The homotetrameric assembly contains two dimers, exhibiting
strong intermonomer interactions within each dimer but more limited ones between
dimers. Each monomeric subunit is partitioned into three structural domains, all
folding according to a mixed alpha/beta motif. Two of these domains are
associated with cofactor binding, while the other is associated with substrate
activation. The catalytic centers containing both thiamin diphosphate and Mg(II)
are located deep in the intermonomer interface within each dimer. Amino acids
important in cofactor binding and likely to participate in catalysis and
substrate activation are identified.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
X.Y.Pei,
K.M.Erixon,
B.F.Luisi,
and
F.J.Leeper
(2010).
Structural insights into the prereaction state of pyruvate decarboxylase from Zymomonas mobilis .
|
| |
Biochemistry,
49,
1727-1736.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Shaanan,
and
D.M.Chipman
(2009).
Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue.
|
| |
FEBS J,
276,
2447-2453.
|
 |
|
|
|
|
 |
N.S.Nemeria,
S.Chakraborty,
A.Balakrishnan,
and
F.Jordan
(2009).
Reaction mechanisms of thiamin diphosphate enzymes: defining states of ionization and tautomerization of the cofactor at individual steps.
|
| |
FEBS J,
276,
2432-3446.
|
 |
|
|
|
|
 |
A.Kaplun,
E.Binshtein,
M.Vyazmensky,
A.Steinmetz,
Z.Barak,
D.M.Chipman,
K.Tittmann,
and
B.Shaanan
(2008).
Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes.
|
| |
Nat Chem Biol,
4,
113-118.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Alstrup Lie,
and
B.Schiøtt
(2008).
A DFT study of solvation effects on the tautomeric equilibrium and catalytic ylide generation of thiamin models.
|
| |
J Comput Chem,
29,
1037-1047.
|
 |
|
|
|
|
 |
T.Werther,
M.Spinka,
K.Tittmann,
A.Schütz,
R.Golbik,
C.Mrestani-Klaus,
G.Hübner,
and
S.König
(2008).
Amino acids allosterically regulate the thiamine diphosphate-dependent alpha-keto acid decarboxylase from Mycobacterium tuberculosis.
|
| |
J Biol Chem,
283,
5344-5354.
|
 |
|
|
|
|
 |
V.I.Bunik,
and
D.Degtyarev
(2008).
Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins.
|
| |
Proteins,
71,
874-890.
|
 |
|
|
|
|
 |
H.Xie,
S.Vucetic,
L.M.Iakoucheva,
C.J.Oldfield,
A.K.Dunker,
Z.Obradovic,
and
V.N.Uversky
(2007).
Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins.
|
| |
J Proteome Res,
6,
1917-1932.
|
 |
|
|
|
|
 |
K.M.Erixon,
C.L.Dabalos,
and
F.J.Leeper
(2007).
Inhibition of pyruvate decarboxylase from Z. mobilis by novel analogues of thiamine pyrophosphate: investigating pyrophosphate mimics.
|
| |
Chem Commun (Camb),
(),
960-962.
|
 |
|
|
|
|
 |
N.Nemeria,
S.Chakraborty,
A.Baykal,
L.G.Korotchkina,
M.S.Patel,
and
F.Jordan
(2007).
The 1',4'-iminopyrimidine tautomer of thiamin diphosphate is poised for catalysis in asymmetric active centers on enzymes.
|
| |
Proc Natl Acad Sci U S A,
104,
78-82.
|
 |
|
|
|
|
 |
S.Kutter,
M.Spinka,
M.H.Koch,
and
S.König
(2007).
The influence of protein concentration on oligomer structure and catalytic function of two pyruvate decarboxylases.
|
| |
Protein J,
26,
585-591.
|
 |
|
|
|
|
 |
G.Malandrinos,
M.Louloudi,
and
N.Hadjiliadis
(2006).
Thiamine models and perspectives on the mechanism of action of thiamine-dependent enzymes.
|
| |
Chem Soc Rev,
35,
684-692.
|
 |
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
P.Arjunan,
M.Sax,
A.Brunskill,
K.Chandrasekhar,
N.Nemeria,
S.Zhang,
F.Jordan,
and
W.Furey
(2006).
A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct.
|
| |
J Biol Chem,
281,
15296-15303.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kutter,
G.Wille,
S.Relle,
M.S.Weiss,
G.Hübner,
and
S.König
(2006).
The crystal structure of pyruvate decarboxylase from Kluyveromyces lactis. Implications for the substrate activation mechanism of this enzyme.
|
| |
FEBS J,
273,
4199-4209.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.A.Smit,
J.E.van Hylckama Vlieg,
W.J.Engels,
L.Meijer,
J.T.Wouters,
and
G.Smit
(2005).
Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation.
|
| |
Appl Environ Microbiol,
71,
303-311.
|
 |
|
|
|
|
 |
S.S.Pang,
R.G.Duggleby,
R.L.Schowen,
and
L.W.Guddat
(2004).
The crystal structures of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate.
|
| |
J Biol Chem,
279,
2242-2253.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.M.Ciszak,
L.G.Korotchkina,
P.M.Dominiak,
S.Sidhu,
and
M.S.Patel
(2003).
Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase.
|
| |
J Biol Chem,
278,
21240-21246.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Zhang,
J.Dai,
Z.Lu,
and
D.Dunaway-Mariano
(2003).
The phosphonopyruvate decarboxylase from Bacteroides fragilis.
|
| |
J Biol Chem,
278,
41302-41308.
|
 |
|
|
|
|
 |
S.S.Pang,
L.W.Guddat,
and
R.G.Duggleby
(2003).
Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase.
|
| |
J Biol Chem,
278,
7639-7644.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.A.Sergienko,
and
F.Jordan
(2002).
New model for activation of yeast pyruvate decarboxylase by substrate consistent with the alternating sites mechanism: demonstration of the existence of two active forms of the enzyme.
|
| |
Biochemistry,
41,
3952-3967.
|
 |
|
|
|
|
 |
J.Wang,
R.Golbik,
B.Seliger,
M.Spinka,
K.Tittmann,
G.Hübner,
and
F.Jordan
(2001).
Consequences of a modified putative substrate-activation site on catalysis by yeast pyruvate decarboxylase.
|
| |
Biochemistry,
40,
1755-1763.
|
 |
|
|
|
|
 |
S.S.Pang,
L.W.Guddat,
and
R.G.Duggleby
(2001).
Crystallization of the catalytic subunit of Saccharomyces cerevisiae acetohydroxyacid synthase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1321-1323.
|
 |
|
|
|
|
 |
A.K.Chang,
P.F.Nixon,
and
R.G.Duggleby
(2000).
Effects of deletions at the carboxyl terminus of Zymomonas mobilis pyruvate decarboxylase on the kinetic properties and substrate specificity.
|
| |
Biochemistry,
39,
9430-9437.
|
 |
|
|
|
|
 |
D.I.Svergun,
M.V.Petoukhov,
M.H.Koch,
and
S.König
(2000).
Crystal versus solution structures of thiamine diphosphate-dependent enzymes.
|
| |
J Biol Chem,
275,
297-302.
|
 |
|
|
|
|
 |
E.A.Sergienko,
J.Wang,
L.Polovnikova,
M.S.Hasson,
M.J.McLeish,
G.L.Kenyon,
and
F.Jordan
(2000).
Spectroscopic detection of transient thiamin diphosphate-bound intermediates on benzoylformate decarboxylase.
|
| |
Biochemistry,
39,
13862-13869.
|
 |
|
|
|
|
 |
G.Lu,
D.Dobritzsch,
S.Baumann,
G.Schneider,
and
S.König
(2000).
The structural basis of substrate activation in yeast pyruvate decarboxylase. A crystallographic and kinetic study.
|
| |
Eur J Biochem,
267,
861-868.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.P.Ward,
and
A.Singh
(2000).
Enzymatic asymmetric synthesis by decarboxylases.
|
| |
Curr Opin Biotechnol,
11,
520-526.
|
 |
|
|
|
|
 |
Y.G.Wu,
A.K.Chang,
P.F.Nixon,
W.Li,
and
R.G.Duggleby
(2000).
Mutagenesis at asp27 of pyruvate decarboxylase from Zymomonas mobilis. Effect on its ability to form acetoin and acetolactate.
|
| |
Eur J Biochem,
267,
6493-6500.
|
 |
|
|
|
|
 |
F.Jordan,
H.Li,
and
A.Brown
(1999).
Remarkable stabilization of zwitterionic intermediates may account for a billion-fold rate acceleration by thiamin diphosphate-dependent decarboxylases.
|
| |
Biochemistry,
38,
6369-6373.
|
 |
|
|
|
|
 |
H.J.Chiu,
J.J.Reddick,
T.P.Begley,
and
S.E.Ealick
(1999).
Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution.
|
| |
Biochemistry,
38,
6460-6470.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Eberhardt,
H.Cederberg,
H.Li,
S.König,
F.Jordan,
and
S.Hohmann
(1999).
Autoregulation of yeast pyruvate decarboxylase gene expression requires the enzyme but not its catalytic activity.
|
| |
Eur J Biochem,
262,
191-201.
|
 |
|
|
|
|
 |
D.Dobritzsch,
S.König,
G.Schneider,
and
G.Lu
(1998).
High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases.
|
| |
J Biol Chem,
273,
20196-20204.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Guo,
D.Zhang,
A.Kahyaoglu,
R.S.Farid,
and
F.Jordan
(1998).
Is a hydrophobic amino acid required to maintain the reactive V conformation of thiamin at the active center of thiamin diphosphate-requiring enzymes? Experimental and computational studies of isoleucine 415 of yeast pyruvate decarboxylase.
|
| |
Biochemistry,
37,
13379-13391.
|
 |
|
|
|
|
 |
F.Jordan,
N.Nemeria,
F.Guo,
I.Baburina,
Y.Gao,
A.Kahyaoglu,
H.Li,
J.Wang,
J.Yi,
J.R.Guest,
and
W.Furey
(1998).
Regulation of thiamin diphosphate-dependent 2-oxo acid decarboxylases by substrate and thiamin diphosphate.Mg(II) - evidence for tertiary and quaternary interactions.
|
| |
Biochim Biophys Acta,
1385,
287-306.
|
 |
|
|
|
|
 |
G.Schenk,
R.G.Duggleby,
and
P.F.Nixon
(1998).
Properties and functions of the thiamin diphosphate dependent enzyme transketolase.
|
| |
Int J Biochem Cell Biol,
30,
1297-1318.
|
 |
|
|
|
|
 |
G.Schneider,
and
Y.Lindqvist
(1998).
Crystallography and mutagenesis of transketolase: mechanistic implications for enzymatic thiamin catalysis.
|
| |
Biochim Biophys Acta,
1385,
387-398.
|
 |
|
|
|
|
 |
I.Baburina,
G.Dikdan,
F.Guo,
G.I.Tous,
B.Root,
and
F.Jordan
(1998).
Reactivity at the substrate activation site of yeast pyruvate decarboxylase: inhibition by distortion of domain interactions.
|
| |
Biochemistry,
37,
1245-1255.
|
 |
|
|
|
|
 |
I.Baburina,
H.Li,
B.Bennion,
W.Furey,
and
F.Jordan
(1998).
Interdomain information transfer during substrate activation of yeast pyruvate decarboxylase: the interaction between cysteine 221 and histidine 92.
|
| |
Biochemistry,
37,
1235-1244.
|
 |
|
|
|
|
 |
J.M.Candy,
and
R.G.Duggleby
(1998).
Structure and properties of pyruvate decarboxylase and site-directed mutagenesis of the Zymomonas mobilis enzyme.
|
| |
Biochim Biophys Acta,
1385,
323-338.
|
 |
|
|
|
|
 |
M.S.Hasson,
A.Muscate,
M.J.McLeish,
L.S.Polovnikova,
J.A.Gerlt,
G.L.Kenyon,
G.A.Petsko,
and
D.Ringe
(1998).
The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
|
| |
Biochemistry,
37,
9918-9930.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Lu,
B.P.Davis,
and
T.W.Jeffries
(1998).
Cloning and characterization of two pyruvate decarboxylase genes from Pichia stipitis CBS 6054.
|
| |
Appl Environ Microbiol,
64,
94-97.
|
 |
|
|
|
|
 |
R.M.Wynn,
J.R.Davie,
J.L.Chuang,
C.D.Cote,
and
D.T.Chuang
(1998).
Impaired assembly of E1 decarboxylase of the branched-chain alpha-ketoacid dehydrogenase complex in type IA maple syrup urine disease.
|
| |
J Biol Chem,
273,
13110-13118.
|
 |
|
|
|
|
 |
S.König,
D.I.Svergun,
V.V.Volkov,
L.A.Feigin,
and
M.H.Koch
(1998).
Small-angle X-ray solution-scattering studies on ligand-induced subunit interactions of the thiamine diphosphate dependent enzyme pyruvate decarboxylase from different organisms.
|
| |
Biochemistry,
37,
5329-5334.
|
 |
|
|
|
|
 |
A.F.Hengeveld,
A.H.Westphal,
and
A.de Kok
(1997).
Expression and characterisation of the homodimeric E1 component of the Azotobacter vinelandii pyruvate dehydrogenase complex.
|
| |
Eur J Biochem,
250,
260-268.
|
 |
|
|
|
|
 |
G.Schenk,
F.J.Leeper,
R.England,
P.F.Nixon,
and
R.G.Duggleby
(1997).
The role of His113 and His114 in pyruvate decarboxylase from Zymomonas mobilis.
|
| |
Eur J Biochem,
248,
63-71.
|
 |
|
|
|
|
 |
K.Ma,
A.Hutchins,
S.J.Sung,
and
M.W.Adams
(1997).
Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase.
|
| |
Proc Natl Acad Sci U S A,
94,
9608-9613.
|
 |
|
|
|
|
 |
M.Killenberg-Jabs,
S.König,
I.Eberhardt,
S.Hohmann,
and
G.Hübner
(1997).
Role of Glu51 for cofactor binding and catalytic activity in pyruvate decarboxylase from yeast studied by site-directed mutagenesis.
|
| |
Biochemistry,
36,
1900-1905.
|
 |
|
|
|
|
 |
M.Vyazmensky,
C.Sella,
Z.Barak,
and
D.M.Chipman
(1996).
Isolation and characterization of subunits of acetohydroxy acid synthase isozyme III and reconstitution of the holoenzyme.
|
| |
Biochemistry,
35,
10339-10346.
|
 |
|
|
|
|
 |
U.Mücke,
T.Wohlfarth,
U.Fiedler,
H.Bäumlein,
K.P.Rücknagel,
and
S.König
(1996).
Pyruvate decarboxylase from Pisum sativum. Properties, nucleotide and amino acid sequences.
|
| |
Eur J Biochem,
237,
373-382.
|
 |
|
|
|
|
 |
H.Bruhn,
M.Pohl,
J.Grötzinger,
and
M.R.Kula
(1995).
The replacement of Trp392 by alanine influences the decarboxylase/carboligase activity and stability of pyruvate decarboxylase from Zymomonas mobilis.
|
| |
Eur J Biochem,
234,
650-655.
|
 |
|
|
|
|
 |
M.S.Hasson,
A.Muscate,
G.T.Henehan,
P.F.Guidinger,
G.A.Petsko,
D.Ringe,
and
G.L.Kenyon
(1995).
Purification and crystallization of benzoylformate decarboxylase.
|
| |
Protein Sci,
4,
955-959.
|
 |
|
|
|
|
 |
U.Mücke,
S.König,
and
G.Hübner
(1995).
Purification and characterisation of pyruvate decarboxylase from pea seeds (Pisum sativum cv. Miko).
|
| |
Biol Chem Hoppe Seyler,
376,
111-117.
|
 |
|
|
|
|
 |
M.Pohl,
J.Grötzinger,
A.Wollmer,
and
M.R.Kula
(1994).
Reversible dissociation and unfolding of pyruvate decarboxylase from Zymomonas mobilis.
|
| |
Eur J Biochem,
224,
651-661.
|
 |
|
|
|
|
 |
Y.A.Muller,
Y.Lindqvist,
W.Furey,
G.E.Schulz,
F.Jordan,
and
G.Schneider
(1993).
A thiamin diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase.
|
| |
Structure,
1,
95.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

