 |
PDBsum entry 1jvb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1jvb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
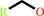 primary alcohol
primary alcohol
|
+
|
 NAD(+)
NAD(+)
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
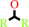 secondary alcohol
secondary alcohol
|
+
|
 NAD(+)
NAD(+)
|
=
|
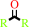 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
318:463-477
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 A resolution.
|
|
L.Esposito,
F.Sica,
C.A.Raia,
A.Giordano,
M.Rossi,
L.Mazzarella,
A.Zagari.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of a medium-chain NAD(H)-dependent alcohol dehydrogenase
(ADH) from an archaeon has been solved by multiwavelength anomalous diffraction,
using a selenomethionine-substituted enzyme. The protein (SsADH), extracted from
the hyperthermophilic organism Sulfolobus solfataricus, is a homo-tetramer with
a crystallographic 222 symmetry. Despite the low level of sequence identity, the
overall fold of the monomer is similar to that of the other homologous ADHs of
known structure. However, a significant difference is the orientation of the
catalytic domain relative to the coenzyme-binding domain that results in a
larger interdomain cleft. At the bottom of this cleft, the catalytic zinc ion is
coordinated tetrahedrally and lacks the zinc-bound water molecule that is
usually found in ADH apoform structures. The fourth coordination position is
indeed occupied by a Glu residue, as found in bacterial tetrameric ADHs. Other
differences are found in the architecture of the substrate pocket whose entrance
is more restricted than in other ADHs. SsADH is the first tetrameric ADH X-ray
structure containing a second zinc ion playing a structural role. This latter
metal ion shows a peculiar coordination, with a glutamic acid residue replacing
one of the four cysteine ligands that are highly conserved throughout the
structural zinc-containing dimeric ADHs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. A stereo view of the SsADH subunit. The C^a trace
is marked with a ball every tenth C^a atom. Figure 1, Figure 3
and Figure 4 were prepared with MOLSCRIPT. [48.]
|
 |
Figure 5.
Figure 5. Ribbon diagram of the SsADH homo-tetramer
generated from crystallographic symmetry. The tetramer is a
dimer of dimers (A/B and C/D). The catalytic zinc ions are
coloured green. The Figure was prepared with MOLSCRIPT[48.] and
Raster3D. [50.]
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
318,
463-477)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.A.Littlechild
(2011).
Thermophilic archaeal enzymes and applications in biocatalysis.
|
| |
Biochem Soc Trans,
39,
155-158.
|
 |
|
|
|
|
 |
B.V.Plapp
(2010).
Conformational changes and catalysis by alcohol dehydrogenase.
|
| |
Arch Biochem Biophys,
493,
3.
|
 |
|
|
|
|
 |
A.Pennacchio,
L.Esposito,
A.Zagari,
M.Rossi,
and
C.A.Raia
(2009).
Role of Tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase.
|
| |
Extremophiles,
13,
751-761.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.N.Marino-Marmolejo,
A.De León-Rodríguez,
A.P.de la Rosa,
and
L.Santos
(2009).
Heterologous Expression and Characterization of an Alcohol Dehydrogenase from the Archeon Thermoplasma acidophilum.
|
| |
Mol Biotechnol,
42,
61-67.
|
 |
|
|
|
|
 |
H.Yanai,
K.Doi,
and
T.Ohshima
(2009).
Sulfolobus tokodaii ST0053 produces a novel thermostable, NAD-dependent medium-chain alcohol dehydrogenase.
|
| |
Appl Environ Microbiol,
75,
1758-1763.
|
 |
|
|
|
|
 |
Q.Bashir,
N.Rashid,
F.Jamil,
T.Imanaka,
and
M.Akhtar
(2009).
Highly thermostable L-threonine dehydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis.
|
| |
J Biochem,
146,
95.
|
 |
|
|
|
|
 |
R.Teufel,
J.W.Kung,
D.Kockelkorn,
B.E.Alber,
and
G.Fuchs
(2009).
3-hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in the Sulfolobales.
|
| |
J Bacteriol,
191,
4572-4581.
|
 |
|
|
|
|
 |
B.Persson,
J.Hedlund,
and
H.Jörnvall
(2008).
Medium- and short-chain dehydrogenase/reductase gene and protein families : the MDR superfamily.
|
| |
Cell Mol Life Sci,
65,
3879-3894.
|
 |
|
|
|
|
 |
E.Goihberg,
O.Dym,
S.Tel-Or,
L.Shimon,
F.Frolow,
M.Peretz,
and
Y.Burstein
(2008).
Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
|
| |
Proteins,
72,
711-719.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Goihberg,
O.Dym,
S.Tel-Or,
I.Levin,
M.Peretz,
and
Y.Burstein
(2007).
A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase.
|
| |
Proteins,
66,
196-204.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Ying,
Y.Wang,
H.R.Badiei,
V.Karanassios,
and
K.Ma
(2007).
Purification and characterization of an iron-containing alcohol dehydrogenase in extremely thermophilic bacterium Thermotoga hypogea.
|
| |
Arch Microbiol,
187,
499-510.
|
 |
|
|
|
|
 |
B.Youn,
R.Camacho,
S.G.Moinuddin,
C.Lee,
L.B.Davin,
N.G.Lewis,
and
C.Kang
(2006).
Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4.
|
| |
Org Biomol Chem,
4,
1687-1697.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.J.Shimon,
E.Goihberg,
M.Peretz,
Y.Burstein,
and
F.Frolow
(2006).
Structure of alcohol dehydrogenase from Entamoeba histolytica.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
541-547.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Atomi
(2005).
Recent progress towards the application of hyperthermophiles and their enzymes.
|
| |
Curr Opin Chem Biol,
9,
166-173.
|
 |
|
|
|
|
 |
K.Miyazono,
Y.Sawano,
and
M.Tanokura
(2005).
Crystal structure and structural stability of acylphosphatase from hyperthermophilic archaeon Pyrococcus horikoshii OT3.
|
| |
Proteins,
61,
196-205.
|
 |
|
|
|
|
 |
R.Willaert,
I.Zegers,
L.Wyns,
and
M.Sleutel
(2005).
Protein crystallization in hydrogel beads.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1280-1288.
|
 |
|
|
|
|
 |
S.Liu,
B.S.Dien,
and
M.A.Cotta
(2005).
Functional expression of bacterial Zymobacter palmae pyruvate decarboxylase gene in Lactococcus lactis.
|
| |
Curr Microbiol,
50,
324-328.
|
 |
|
|
|
|
 |
S.Watanabe,
T.Kodaki,
and
K.Makino
(2005).
Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc.
|
| |
J Biol Chem,
280,
10340-10349.
|
 |
|
|
|
|
 |
Y.Papanikolau,
I.Tsigos,
M.Papadovasilaki,
V.Bouriotis,
and
K.Petratos
(2005).
Crystallization and preliminary X-ray diffraction studies of an alcohol dehydrogenase from the Antarctic psychrophile Moraxella sp. TAE123.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
246-248.
|
 |
|
|
|
|
 |
I.Levin,
G.Meiri,
M.Peretz,
Y.Burstein,
and
F.Frolow
(2004).
The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol.
|
| |
Protein Sci,
13,
1547-1556.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.J.Kim,
M.R.Kim,
D.L.Bedgar,
S.G.Moinuddin,
C.L.Cardenas,
L.B.Davin,
C.Kang,
and
N.G.Lewis
(2004).
Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis.
|
| |
Proc Natl Acad Sci U S A,
101,
1455-1460.
|
 |
|
|
|
|
 |
E.Occhipinti,
P.L.Martelli,
F.Spinozzi,
F.Corsi,
C.Formantici,
L.Molteni,
H.Amenitsch,
P.Mariani,
P.Tortora,
and
R.Casadio
(2003).
3D structure of Sulfolobus solfataricus carboxypeptidase developed by molecular modeling is confirmed by site-directed mutagenesis and small angle X-ray scattering.
|
| |
Biophys J,
85,
1165-1175.
|
 |
|
|
|
|
 |
E.T.Powers,
and
D.L.Powers
(2003).
A perspective on mechanisms of protein tetramer formation.
|
| |
Biophys J,
85,
3587-3599.
|
 |
|
|
|
|
 |
G.Fiorentino,
R.Cannio,
M.Rossi,
and
S.Bartolucci
(2003).
Transcriptional regulation of the gene encoding an alcohol dehydrogenase in the archaeon Sulfolobus solfataricus involves multiple factors and control elements.
|
| |
J Bacteriol,
185,
3926-3934.
|
 |
|
|
|
|
 |
H.Radianingtyas,
and
P.C.Wright
(2003).
Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria.
|
| |
FEMS Microbiol Rev,
27,
593-616.
|
 |
|
|
|
|
 |
L.Esposito,
I.Bruno,
F.Sica,
C.A.Raia,
A.Giordano,
M.Rossi,
L.Mazzarella,
and
A.Zagari
(2003).
Crystal structure of a ternary complex of the alcohol dehydrogenase from Sulfolobus solfataricus.
|
| |
Biochemistry,
42,
14397-14407.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Kleifeld,
S.P.Shi,
R.Zarivach,
M.Eisenstein,
and
I.Sagi
(2003).
The conserved Glu-60 residue in Thermoanaerobacter brockii alcohol dehydrogenase is not essential for catalysis.
|
| |
Protein Sci,
12,
468-479.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

