 |
PDBsum entry 1jkf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.5.1.4
- inositol-3-phosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose 6-phosphate = 1D-myo-inositol 3-phosphate
|
 |
 |
 |
 |
 |
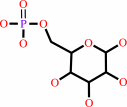 D-glucose 6-phosphate
D-glucose 6-phosphate
|
=
|
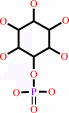 1D-myo-inositol 3-phosphate
1D-myo-inositol 3-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
NAD(+)
|
 |
 |
 |
 |
 |
NAD(+)
Bound ligand (Het Group name =
NAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:9484-9491
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase.
|
|
A.J.Stein,
J.H.Geiger.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
1-l-myo-Inositol-1-phosphate synthase catalyzes the conversion of d-glucose
6-phosphate to 1-l-myo-inositol-1-phosphate (MIP), the first and rate-limiting
step in the biosynthesis of all inositol-containing compounds. It involves an
oxidation, intramolecular aldol cyclization, and reduction. We have determined
the first crystal structure of MIP synthase. We present structures of both the
NAD-bound enzyme and the enzyme bound to an inhibitor,
2-deoxy-glucitol-6-phosphate. While 58 amino acids are disordered in the unbound
form of the enzyme in the vicinity of the active site, the inhibitor nucleates
the folding of this domain in a striking example of induced fit, serving to
completely encapsulate it within the enzyme. Three helices and a long
beta-strand are formed in this process. We postulate a mechanism for the
conversion based on the structure of the inhibitor-bound complex.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Ribbon model of the MIP synthase tetramer in the
absence of inhibitor. This and all other structure figures were
made using the program RIBBONS (55). The top two monomers are
colored red and blue, make up one asymmetric unit, and are
related by a noncrystallographic 2-fold axis that is roughly in
the plane of the page. The bottom two monomers are green and are
related to the top two monomers by a crystallographic 2-fold
axis running roughly perpendicular to the page.
|
 |
Figure 6.
Fig. 6. Proposed mechanism for the transformation
catalyzed by MIP synthase.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
9484-9491)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.S.Seelan,
J.Lakshmanan,
M.F.Casanova,
and
R.N.Parthasarathy
(2009).
Identification of myo-inositol-3-phosphate synthase isoforms: characterization, expression, and putative role of a 16-kDa gamma(c) isoform.
|
| |
J Biol Chem,
284,
9443-9457.
|
 |
|
|
|
|
 |
F.Nieuwmeyer,
K.van der Voort Maarschalk,
and
H.Vromans
(2008).
Lactose contaminant as steroid degradation enhancer.
|
| |
Pharm Res,
25,
2666-2673.
|
 |
|
|
|
|
 |
P.M.Flatt,
and
T.Mahmud
(2007).
Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds.
|
| |
Nat Prod Rep,
24,
358-392.
|
 |
|
|
|
|
 |
A.Chatterjee,
K.G.Dastidar,
S.Maitra,
A.Das-Chatterjee,
H.Dihazi,
K.Eschrich,
and
A.L.Majumder
(2006).
sll1981, an acetolactate synthase homologue of Synechocystis sp. PCC6803, functions as L-myo-inositol 1-phosphate synthase.
|
| |
Planta,
224,
367-379.
|
 |
|
|
|
|
 |
F.Movahedzadeh,
D.A.Smith,
R.A.Norman,
P.Dinadayala,
J.Murray-Rust,
D.G.Russell,
S.L.Kendall,
S.C.Rison,
M.S.McAlister,
G.J.Bancroft,
N.Q.McDonald,
M.Daffe,
Y.Av-Gay,
and
N.G.Stoker
(2004).
The Mycobacterium tuberculosis ino1 gene is essential for growth and virulence.
|
| |
Mol Microbiol,
51,
1003-1014.
|
 |
|
|
|
|
 |
X.Jin,
and
J.H.Geiger
(2003).
Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1154-1164.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

