 |
PDBsum entry 1g6t

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.19
- 3-phosphoshikimate 1-carboxyvinyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-phosphoshikimate + phosphoenolpyruvate = 5-O-(1-carboxyvinyl)-3- phosphoshikimate + phosphate
|
 |
 |
 |
 |
 |
3-phosphoshikimate
|
+
|
phosphoenolpyruvate
Bound ligand (Het Group name = )
matches with 52.94% similarity
|
=
|
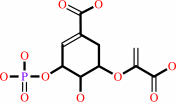 5-O-(1-carboxyvinyl)-3- phosphoshikimate
5-O-(1-carboxyvinyl)-3- phosphoshikimate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
98:1376-1380
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
|
|
E.Schönbrunn,
S.Eschenburg,
W.A.Shuttleworth,
J.V.Schloss,
N.Amrhein,
J.N.Evans,
W.Kabsch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Biosynthesis of aromatic amino acids in plants, many bacteria, and microbes
relies on the enzyme 5-enolpyruvylshikimate 3-phosphate (EPSP) synthase, a prime
target for drugs and herbicides. We have identified the interaction of EPSP
synthase with one of its two substrates (shikimate 3-phosphate) and with the
widely used herbicide glyphosate by x-ray crystallography. The two-domain enzyme
closes on ligand binding, thereby forming the active site in the interdomain
cleft. Glyphosate appears to occupy the binding site of the second substrate of
EPSP synthase (phosphoenol pyruvate), mimicking an intermediate state of the
ternary enzyme.substrates complex. The elucidation of the active site of EPSP
synthase and especially of the binding pattern of glyphosate provides a valuable
roadmap for engineering new herbicides and herbicide-resistant crops, as well as
new antibiotic and antiparasitic drugs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Cartoon of EPSP synthase in the open and closed
conformation. Top domain, residues 20-240; bottom domain,
residues 1-19 plus 241-427. (a) Unliganded state (open) as
reconstructed from the deposited  -carbon
atoms (Protein Data Bank entry code 1EPS). (b) Liganded state
(closed). S3P and glyphosate are shown as ball-and-stick models
in green and magenta, respectively. Drawn with BOBSCRIPT (24). -carbon
atoms (Protein Data Bank entry code 1EPS). (b) Liganded state
(closed). S3P and glyphosate are shown as ball-and-stick models
in green and magenta, respectively. Drawn with BOBSCRIPT (24).
|
 |
Figure 3.
Fig. 3. Schematic representation of ligand binding in the
EPSP synthase·S3P·glyphosate complex. Ligands are
drawn in bold lines. Dashed lines indicate hydrogen bonds and
ionic interactions. Strictly conserved residues are highlighted
by bold labels. Protein atoms are labeled according to the
Protein Data Bank nomenclature. Circled labels W1 to W4
designate solvent molecules. Hydrophobic interactions between
S3P and Tyr-200 are omitted.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Haghani,
K.Khajeh,
A.H.Salmanian,
B.Ranjbar,
and
S.Bakhtiyari
(2011).
Acid-Induced Formation of Molten Globule States in the Wild Type Escherichia coli 5-Enolpyruvylshikimate 3-Phosphate Synthase and its Three Mutated Forms: G96A, A183T and G96A/A183T.
|
| |
Protein J,
30,
132-137.
|
 |
|
|
|
|
 |
S.B.Powles,
and
Q.Yu
(2010).
Evolution in action: plants resistant to herbicides.
|
| |
Annu Rev Plant Biol,
61,
317-347.
|
 |
|
|
|
|
 |
Y.S.Tian,
A.S.Xiong,
J.Xu,
W.Zhao,
F.Gao,
X.Y.Fu,
H.Xu,
J.L.Zheng,
R.H.Peng,
and
Q.H.Yao
(2010).
Isolation from Ochrobactrum anthropi of a novel class II 5-enopyruvylshikimate-3-phosphate synthase with high tolerance to glyphosate.
|
| |
Appl Environ Microbiol,
76,
6001-6005.
|
 |
|
|
|
|
 |
G.B.Barcellos,
R.A.Caceres,
and
W.F.de Azevedo
(2009).
Structural studies of shikimate dehydrogenase from Bacillus anthracis complexed with cofactor NADP.
|
| |
J Mol Model,
15,
147-155.
|
 |
|
|
|
|
 |
T.Funke,
Y.Yang,
H.Han,
M.Healy-Fried,
S.Olesen,
A.Becker,
and
E.Schönbrunn
(2009).
Structural basis of glyphosate resistance resulting from the double mutation Thr97 -> Ile and Pro101 -> Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli.
|
| |
J Biol Chem,
284,
9854-9860.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.J.Vande Berg,
P.E.Hammer,
B.L.Chun,
L.C.Schouten,
B.Carr,
R.Guo,
C.Peters,
T.K.Hinson,
V.Beilinson,
A.Shekita,
R.Deter,
Z.Chen,
V.Samoylov,
C.T.Bryant,
M.E.Stauffer,
T.Eberle,
D.J.Moellenbeck,
N.B.Carozzi,
M.G.Koziel,
and
N.B.Duck
(2008).
Characterization and plant expression of a glyphosate-tolerant enolpyruvylshikimate phosphate synthase.
|
| |
Pest Manag Sci,
64,
340-345.
|
 |
|
|
|
|
 |
A.Perez-Jones,
K.W.Park,
N.Polge,
J.Colquhoun,
and
C.A.Mallory-Smith
(2007).
Investigating the mechanisms of glyphosate resistance in Lolium multiflorum.
|
| |
Planta,
226,
395-404.
|
 |
|
|
|
|
 |
C.H.Huangfu,
X.L.Song,
S.Qiang,
and
H.J.Zhang
(2007).
Response of wild Brassica juncea populations to glyphosate.
|
| |
Pest Manag Sci,
63,
1133-1140.
|
 |
|
|
|
|
 |
M.Hoff,
D.Y.Son,
M.Gubesch,
K.Ahn,
S.I.Lee,
S.Vieths,
R.E.Goodman,
B.K.Ballmer-Weber,
and
G.A.Bannon
(2007).
Serum testing of genetically modified soybeans with special emphasis on potential allergenicity of the heterologous protein CP4 EPSPS.
|
| |
Mol Nutr Food Res,
51,
946-955.
|
 |
|
|
|
|
 |
Q.Yu,
A.Cairns,
and
S.Powles
(2007).
Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype.
|
| |
Planta,
225,
499-513.
|
 |
|
|
|
|
 |
M.D'Alessandro,
M.Held,
Y.Triponez,
and
T.C.Turlings
(2006).
The role of indole and other shikimic acid derived maize volatiles in the attraction of two parasitic wasps.
|
| |
J Chem Ecol,
32,
2733-2748.
|
 |
|
|
|
|
 |
T.Funke,
H.Han,
M.L.Healy-Fried,
M.Fischer,
and
E.Schönbrunn
(2006).
Molecular basis for the herbicide resistance of Roundup Ready crops.
|
| |
Proc Natl Acad Sci U S A,
103,
13010-13015.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Richard,
S.Moslemi,
H.Sipahutar,
N.Benachour,
and
G.E.Seralini
(2005).
Differential effects of glyphosate and roundup on human placental cells and aromatase.
|
| |
Environ Health Perspect,
113,
716-720.
|
 |
|
|
|
|
 |
Y.C.Sun,
Y.C.Chen,
Z.X.Tian,
F.M.Li,
X.Y.Wang,
J.Zhang,
Z.L.Xiao,
M.Lin,
N.Gilmartin,
D.N.Dowling,
and
Y.P.Wang
(2005).
Novel AroA with high tolerance to glyphosate, encoded by a gene of Pseudomonas putida 4G-1 isolated from an extremely polluted environment in China.
|
| |
Appl Environ Microbiol,
71,
4771-4776.
|
 |
|
|
|
|
 |
A.M.Thomas,
C.Ginj,
I.Jelesarov,
N.Amrhein,
and
P.Macheroux
(2004).
Role of K22 and R120 in the covalent binding of the antibiotic fosfomycin and the substrate-induced conformational change in UDP-N-acetylglucosamine enolpyruvyl transferase.
|
| |
Eur J Biochem,
271,
2682-2690.
|
 |
|
|
|
|
 |
C.M.Viola,
V.Saridakis,
and
D.Christendat
(2004).
Crystal structure of chorismate synthase from Aquifex aeolicus reveals a novel beta alpha beta sandwich topology.
|
| |
Proteins,
54,
166-169.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Park,
J.L.Hilsenbeck,
H.J.Kim,
W.A.Shuttleworth,
Y.H.Park,
J.N.Evans,
and
C.Kang
(2004).
Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
|
| |
Mol Microbiol,
51,
963-971.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.K.Padyana,
and
S.K.Burley
(2003).
Crystal structure of shikimate 5-dehydrogenase (SDH) bound to NADP: insights into function and evolution.
|
| |
Structure,
11,
1005-1013.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Vogan
(2003).
Shikimate dehydrogenase structure reveals novel fold.
|
| |
Structure,
11,
902-903.
|
 |
|
|
|
|
 |
H.J.Ahn,
J.K.Yang,
B.I.Lee,
H.J.Yoon,
H.W.Kim,
and
S.W.Suh
(2003).
Crystallization and preliminary X-ray crystallographic studies of chorismate synthase from Helicobacter pylori.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
569-571.
|
 |
|
|
|
|
 |
M.He,
Y.F.Nie,
and
P.Xu
(2003).
A T42M substitution in bacterial 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) generates enzymes with increased resistance to glyphosate.
|
| |
Biosci Biotechnol Biochem,
67,
1405-1409.
|
 |
|
|
|
|
 |
S.Ye,
F.Von Delft,
A.Brooun,
M.W.Knuth,
R.V.Swanson,
and
D.E.McRee
(2003).
The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode.
|
| |
J Bacteriol,
185,
4144-4151.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.F.Alibhai,
and
W.C.Stallings
(2001).
Closing down on glyphosate inhibition--with a new structure for drug discovery.
|
| |
Proc Natl Acad Sci U S A,
98,
2944-2946.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

