 |
PDBsum entry 1g0r

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (rmla). Thymidine/glucose- 1-phosphate complex.
|
|
Structure:
|
 |
Glucose-1-phosphate thymidylyltransferase. Chain: a, b, c, d, e, f, g, h. Synonym: glucose-1-phosphate thymidylyltransferase (rmla). Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
1.87Å
|
R-factor:
|
0.151
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
W.Blankenfeldt,M.Asuncion,J.S.Lam,J.H.Naismith
|
Key ref:

|
 |
W.Blankenfeldt
et al.
(2000).
The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J,
19,
6652-6663.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Oct-00
|
Release date:
|
27-Dec-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9HU22
(Q9HU22_PSEAE) -
Glucose-1-phosphate thymidylyltransferase from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
293 a.a.
292 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.24
- glucose-1-phosphate thymidylyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
6-Deoxyhexose Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dTTP + alpha-D-glucose 1-phosphate + H+ = dTDP-alpha-D-glucose + diphosphate
|
 |
 |
 |
 |
 |
dTTP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
alpha-D-glucose 1-phosphate
Bound ligand (Het Group name = )
matches with 58.62% similarity
|
+
|
H(+)
|
=
|
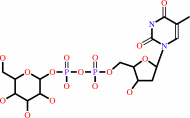 dTDP-alpha-D-glucose
dTDP-alpha-D-glucose
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
19:6652-6663
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
|
|
W.Blankenfeldt,
M.Asuncion,
J.S.Lam,
J.H.Naismith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The synthesis of deoxy-thymidine di-phosphate (dTDP)-L-rhamnose, an important
component of the cell wall of many microorganisms, is a target for therapeutic
intervention. The first enzyme in the dTDP-L-rhamnose biosynthetic pathway is
glucose-1-phosphate thymidylyltransferase (RmlA). RmlA is inhibited by
dTDP-L-rhamnose thereby regulating L-rhamnose production in bacteria. The
structure of Pseudomonas aeruginosa RmlA has been solved to 1.66 A resolution.
RmlA is a homotetramer, with the monomer consisting of three functional
subdomains. The sugar binding and dimerization subdomains are unique to
RmlA-like enzymes. The sequence of the core subdomain is found not only in sugar
nucleotidyltransferases but also in other nucleotidyltransferases. The
structures of five distinct enzyme substrate- product complexes reveal the
enzyme mechanism that involves precise positioning of the nucleophile and
activation of the electrophile. All the key residues are within the core
subdomain, suggesting that the basic mechanism is found in many
nucleotidyltransferases. The dTDP-L-rhamnose complex identifies how the protein
is controlled by its natural inhibitor. This work provides a platform for the
design of novel drugs against pathogenic bacteria.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 (A) The mechanism of the reaction catalysed by RmlA.
(B) The distinct chemical groups that form the ternary complex
with the protein.
|
 |
Figure 2.
Figure 2 (A) Stereo ribbon diagram of the RmlA monomer with
location of secondary structure elements. The different colours
denote the three subdomains. Yellow is the core binding
subdomain, light blue is the sugar-binding subdomain and magenta
the dimerization subdomain. The  character
represents a 3[10] helix, and character
represents a 3[10] helix, and  and and
 have
their normal meaning. Secondary structure was assigned with DSSP
(Kabsch and Sander, 1983). (B) A ribbon representation of the
RmlA tetramer. The monomers are coloured red, monomer A; blue,
monomer B; yellow, monomer A'; and light blue, monomer B'. G-1-P
(black) and dTTP (green) are shown at the active sites in
ball-and-stick format. dTDP–L-rhamnose (magenta) is shown in
the secondary binding sites, again as a ball-and-stick diagram.
(C) Same as (B), rotated by 90° around the y-axis. All
molecular representations are prepared with BOBSCRIPT (Esnouf,
1997) through the GL_RENDER interface (L.Esser and
J.Deisenhofer, unpublished data) and were rendered with
POV-Ray™. have
their normal meaning. Secondary structure was assigned with DSSP
(Kabsch and Sander, 1983). (B) A ribbon representation of the
RmlA tetramer. The monomers are coloured red, monomer A; blue,
monomer B; yellow, monomer A'; and light blue, monomer B'. G-1-P
(black) and dTTP (green) are shown at the active sites in
ball-and-stick format. dTDP–L-rhamnose (magenta) is shown in
the secondary binding sites, again as a ball-and-stick diagram.
(C) Same as (B), rotated by 90° around the y-axis. All
molecular representations are prepared with BOBSCRIPT (Esnouf,
1997) through the GL_RENDER interface (L.Esser and
J.Deisenhofer, unpublished data) and were rendered with
POV-Ray™.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2000,
19,
6652-6663)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.S.Reis,
A.G.Pereira,
B.C.Neves,
and
D.M.Freire
(2011).
Gene regulation of rhamnolipid production in Pseudomonas aeruginosa--a review.
|
| |
Bioresour Technol,
102,
6377-6384.
|
 |
|
|
|
|
 |
H.Denton,
S.Fyffe,
and
T.K.Smith
(2010).
GDP-mannose pyrophosphorylase is essential in the bloodstream form of Trypanosoma brucei.
|
| |
Biochem J,
425,
603-614.
|
 |
|
|
|
|
 |
H.Kim,
J.Choi,
T.Kim,
N.K.Lokanath,
S.C.Ha,
S.W.Suh,
H.Y.Hwang,
and
K.K.Kim
(2010).
Structural basis for the reaction mechanism of UDP-glucose pyrophosphorylase.
|
| |
Mol Cells,
29,
397-405.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Simkhada,
T.J.Oh,
E.M.Kim,
J.C.Yoo,
and
J.K.Sohng
(2009).
Cloning and characterization of CalS7 from Micromonospora echinospora sp. calichensis as a glucose-1-phosphate nucleotidyltransferase.
|
| |
Biotechnol Lett,
31,
147-153.
|
 |
|
|
|
|
 |
C.J.Zea,
G.Camci-Unal,
and
N.L.Pohl
(2008).
Thermodynamics of binding of divalent magnesium and manganese to uridine phosphates: implications for diabetes-related hypomagnesaemia and carbohydrate biocatalysis.
|
| |
Chem Cent J,
2,
15.
|
 |
|
|
|
|
 |
D.J.McNally,
I.C.Schoenhofen,
R.S.Houliston,
N.H.Khieu,
D.M.Whitfield,
S.M.Logan,
H.C.Jarrell,
and
J.R.Brisson
(2008).
CMP-pseudaminic acid is a natural potent inhibitor of PseB, the first enzyme of the pseudaminic acid pathway in Campylobacter jejuni and Helicobacter pylori.
|
| |
ChemMedChem,
3,
55-59.
|
 |
|
|
|
|
 |
M.P.Huestis,
G.A.Aish,
J.P.Hui,
E.C.Soo,
and
D.L.Jakeman
(2008).
Lipophilic sugar nucleotide synthesis by structure-based design of nucleotidylyltransferase substrates.
|
| |
Org Biomol Chem,
6,
477-484.
|
 |
|
|
|
|
 |
R.Moretti,
and
J.S.Thorson
(2008).
A comparison of sugar indicators enables a universal high-throughput sugar-1-phosphate nucleotidyltransferase assay.
|
| |
Anal Biochem,
377,
251-258.
|
 |
|
|
|
|
 |
S.K.Hwang,
Y.Nagai,
D.Kim,
and
T.W.Okita
(2008).
Direct appraisal of the potato tuber ADP-glucose pyrophosphorylase large subunit in enzyme function by study of a novel mutant form.
|
| |
J Biol Chem,
283,
6640-6647.
|
 |
|
|
|
|
 |
S.L.Rivera,
E.Vargas,
M.I.Ramírez-Díaz,
J.Campos-García,
and
C.Cervantes
(2008).
Genes related to chromate resistance by Pseudomonas aeruginosa PAO1.
|
| |
Antonie Van Leeuwenhoek,
94,
299-305.
|
 |
|
|
|
|
 |
C.Dong,
L.L.Major,
V.Srikannathasan,
J.C.Errey,
M.F.Giraud,
J.S.Lam,
M.Graninger,
P.Messner,
M.R.McNeil,
R.A.Field,
C.Whitfield,
and
J.H.Naismith
(2007).
RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation.
|
| |
J Mol Biol,
365,
146-159.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Schäffer,
R.Novotny,
S.Küpcü,
S.Zayni,
A.Scheberl,
J.Friedmann,
U.B.Sleytr,
and
P.Messner
(2007).
Novel biocatalysts based on S-layer self-assembly of Geobacillus stearothermophilus NRS 2004/3a: a nanobiotechnological approach.
|
| |
Small,
3,
1549-1559.
|
 |
|
|
|
|
 |
D.Aragão,
A.M.Fialho,
A.R.Marques,
E.P.Mitchell,
I.Sá-Correia,
and
C.Frazão
(2007).
The complex of Sphingomonas elodea ATCC 31461 glucose-1-phosphate uridylyltransferase with glucose-1-phosphate reveals a novel quaternary structure, unique among nucleoside diphosphate-sugar pyrophosphorylase members.
|
| |
J Bacteriol,
189,
4520-4528.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Maruyama,
Y.Nishitani,
T.Nonaka,
A.Kita,
T.A.Fukami,
T.Mio,
H.Yamada-Okabe,
T.Yamada-Okabe,
and
K.Miki
(2007).
Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans and catalytic reaction mechanism.
|
| |
J Biol Chem,
282,
17221-17230.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Mochalkin,
S.Lightle,
Y.Zhu,
J.F.Ohren,
C.Spessard,
N.Y.Chirgadze,
C.Banotai,
M.Melnick,
and
L.McDowell
(2007).
Characterization of substrate binding and catalysis in the potential antibacterial target N-acetylglucosamine-1-phosphate uridyltransferase (GlmU).
|
| |
Protein Sci,
16,
2657-2666.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
and
H.M.Holden
(2007).
The molecular architecture of glucose-1-phosphate uridylyltransferase.
|
| |
Protein Sci,
16,
432-440.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Thoden,
and
H.M.Holden
(2007).
Active site geometry of glucose-1-phosphate uridylyltransferase.
|
| |
Protein Sci,
16,
1379-1388.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Steiner,
A.C.Lamerz,
P.Hess,
C.Breithaupt,
S.Krapp,
G.Bourenkov,
R.Huber,
R.Gerardy-Schahn,
and
U.Jacob
(2007).
Open and closed structures of the UDP-glucose pyrophosphorylase from Leishmania major.
|
| |
J Biol Chem,
282,
13003-13010.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.M.Bejar,
X.Jin,
M.A.Ballicora,
and
J.Preiss
(2006).
Molecular architecture of the glucose 1-phosphate site in ADP-glucose pyrophosphorylases.
|
| |
J Biol Chem,
281,
40473-40484.
|
 |
|
|
|
|
 |
D.Aragão,
A.R.Marques,
C.Frazão,
F.J.Enguita,
M.A.Carrondo,
A.M.Fialho,
I.Sá-Correia,
and
E.P.Mitchell
(2006).
Cloning, expression, purification, crystallization and preliminary structure determination of glucose-1-phosphate uridylyltransferase (UgpG) from Sphingomonas elodea ATCC 31461 bound to glucose-1-phosphate.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
930-934.
|
 |
|
|
|
|
 |
E.Silva,
A.R.Marques,
A.M.Fialho,
A.T.Granja,
and
I.Sá-Correia
(2005).
Proteins encoded by Sphingomonas elodea ATCC 31461 rmlA and ugpG genes, involved in gellan gum biosynthesis, exhibit both dTDP- and UDP-glucose pyrophosphorylase activities.
|
| |
Appl Environ Microbiol,
71,
4703-4712.
|
 |
|
|
|
|
 |
I.T.Kudva,
R.W.Griffin,
J.M.Garren,
S.B.Calderwood,
and
M.John
(2005).
Identification of a protein subset of the anthrax spore immunome in humans immunized with the anthrax vaccine adsorbed preparation.
|
| |
Infect Immun,
73,
5685-5696.
|
 |
|
|
|
|
 |
J.Bae,
K.H.Kim,
D.Kim,
Y.Choi,
J.S.Kim,
S.Koh,
S.I.Hong,
and
D.S.Lee
(2005).
A practical enzymatic synthesis of UDP sugars and NDP glucoses.
|
| |
Chembiochem,
6,
1963-1966.
|
 |
|
|
|
|
 |
J.R.Cupp-Vickery,
R.Y.Igarashi,
and
C.R.Meyer
(2005).
Preliminary crystallographic analysis of ADP-glucose pyrophosphorylase from Agrobacterium tumefaciens.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
266-268.
|
 |
|
|
|
|
 |
K.Usui,
S.Katayama,
M.Kanamori-Katayama,
C.Ogawa,
C.Kai,
M.Okada,
J.Kawai,
T.Arakawa,
P.Carninci,
M.Itoh,
K.Takio,
M.Miyano,
S.Kidoaki,
T.Matsuda,
Y.Hayashizaki,
and
H.Suzuki
(2005).
Protein-protein interactions of the hyperthermophilic archaeon Pyrococcus horikoshii OT3.
|
| |
Genome Biol,
6,
R98.
|
 |
|
|
|
|
 |
M.A.Ballicora,
J.R.Dubay,
C.H.Devillers,
and
J.Preiss
(2005).
Resurrecting the ancestral enzymatic role of a modulatory subunit.
|
| |
J Biol Chem,
280,
10189-10195.
|
 |
|
|
|
|
 |
M.A.Perugini,
M.D.Griffin,
B.J.Smith,
L.E.Webb,
A.J.Davis,
E.Handman,
and
J.A.Gerrard
(2005).
Insight into the self-association of key enzymes from pathogenic species.
|
| |
Eur Biophys J,
34,
469-476.
|
 |
|
|
|
|
 |
X.Jin,
M.A.Ballicora,
J.Preiss,
and
J.H.Geiger
(2005).
Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
|
| |
EMBO J,
24,
694-704.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Zhang,
M.Tsujimura,
J.Akutsu,
M.Sasaki,
H.Tajima,
and
Y.Kawarabayasi
(2005).
Identification of an extremely thermostable enzyme with dual sugar-1-phosphate nucleotidylyltransferase activities from an acidothermophilic archaeon, Sulfolobus tokodaii strain 7.
|
| |
J Biol Chem,
280,
9698-9705.
|
 |
|
|
|
|
 |
C.Kent,
P.Gee,
S.Y.Lee,
X.Bian,
and
J.C.Fenno
(2004).
A CDP-choline pathway for phosphatidylcholine biosynthesis in Treponema denticola.
|
| |
Mol Microbiol,
51,
471-481.
|
 |
|
|
|
|
 |
J.S.Thorson,
W.A.Barton,
D.Hoffmeister,
C.Albermann,
and
D.B.Nikolov
(2004).
Structure-based enzyme engineering and its impact on in vitro glycorandomization.
|
| |
Chembiochem,
5,
16-25.
|
 |
|
|
|
|
 |
N.M.Koropatkin,
and
H.M.Holden
(2004).
Molecular structure of alpha-D-glucose-1-phosphate cytidylyltransferase from Salmonella typhi.
|
| |
J Biol Chem,
279,
44023-44029.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Matte,
J.Sivaraman,
I.Ekiel,
K.Gehring,
Z.Jia,
and
M.Cygler
(2003).
Contribution of structural genomics to understanding the biology of Escherichia coli.
|
| |
J Bacteriol,
185,
3994-4002.
|
 |
|
|
|
|
 |
B.A.Wolucka,
and
M.Van Montagu
(2003).
GDP-mannose 3',5'-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants.
|
| |
J Biol Chem,
278,
47483-47490.
|
 |
|
|
|
|
 |
J.B.Frueauf,
M.A.Ballicora,
and
J.Preiss
(2003).
ADP-glucose pyrophosphorylase from potato tuber: site-directed mutagenesis of homologous aspartic acid residues in the small and large subunits.
|
| |
Plant J,
33,
503-511.
|
 |
|
|
|
|
 |
J.Liu,
and
A.Mushegian
(2003).
Three monophyletic superfamilies account for the majority of the known glycosyltransferases.
|
| |
Protein Sci,
12,
1418-1431.
|
 |
|
|
|
|
 |
M.A.Ballicora,
A.A.Iglesias,
and
J.Preiss
(2003).
ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis.
|
| |
Microbiol Mol Biol Rev,
67,
213.
|
 |
|
|
|
|
 |
P.Crevillén,
M.A.Ballicora,
A.Mérida,
J.Preiss,
and
J.M.Romero
(2003).
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct kinetic and regulatory properties to the heterotetrameric enzyme.
|
| |
J Biol Chem,
278,
28508-28515.
|
 |
|
|
|
|
 |
A.B.Merkel,
G.K.Temple,
M.D.Burkart,
H.C.Losey,
K.Beis,
C.T.Walsh,
and
J.H.Naismith
(2002).
Purification, crystallization and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis, the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1226-1228.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Y.Kwak,
Y.M.Zhang,
M.Yun,
R.J.Heath,
C.O.Rock,
S.Jackowski,
and
H.W.Park
(2002).
Structure and mechanism of CTP:phosphocholine cytidylyltransferase (LicC) from Streptococcus pneumoniae.
|
| |
J Biol Chem,
277,
4343-4350.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Sivaraman,
V.Sauvé,
A.Matte,
and
M.Cygler
(2002).
Crystal structure of Escherichia coli glucose-1-phosphate thymidylyltransferase (RffH) complexed with dTTP and Mg2+.
|
| |
J Biol Chem,
277,
44214-44219.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.A.Barton,
J.B.Biggins,
J.Jiang,
J.S.Thorson,
and
D.B.Nikolov
(2002).
Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering.
|
| |
Proc Natl Acad Sci U S A,
99,
13397-13402.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.M.He,
and
H.W.Liu
(2002).
Formation of unusual sugars: mechanistic studies and biosynthetic applications.
|
| |
Annu Rev Biochem,
71,
701-754.
|
 |
|
|
|
|
 |
C.O.Rock,
R.J.Heath,
H.W.Park,
and
S.Jackowski
(2001).
The licC gene of Streptococcus pneumoniae encodes a CTP:phosphocholine cytidylyltransferase.
|
| |
J Bacteriol,
183,
4927-4931.
|
 |
|
|
|
|
 |
C.Peneff,
P.Ferrari,
V.Charrier,
Y.Taburet,
C.Monnier,
V.Zamboni,
J.Winter,
M.Harnois,
F.Fassy,
and
Y.Bourne
(2001).
Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture.
|
| |
EMBO J,
20,
6191-6202.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.B.Frueauf,
M.A.Ballicora,
and
J.Preiss
(2001).
Aspartate residue 142 is important for catalysis by ADP-glucose pyrophosphorylase from Escherichia coli.
|
| |
J Biol Chem,
276,
46319-46325.
|
 |
|
|
|
|
 |
M.F.Giraud,
and
J.H.Naismith
(2000).
The rhamnose pathway.
|
| |
Curr Opin Struct Biol,
10,
687-696.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

