 |
PDBsum entry 2v0h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.3.1.157
- glucosamine-1-phosphate N-acetyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
UDP-N-acetylglucosamine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucosamine 1-phosphate + acetyl-CoA = N-acetyl-alpha-D- glucosamine 1-phosphate + CoA + H+
|
 |
 |
 |
 |
 |
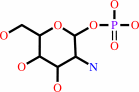 alpha-D-glucosamine 1-phosphate
alpha-D-glucosamine 1-phosphate
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
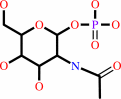 N-acetyl-alpha-D- glucosamine 1-phosphate
N-acetyl-alpha-D- glucosamine 1-phosphate
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.7.23
- UDP-N-acetylglucosamine diphosphorylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-acetyl-alpha-D-glucosamine 1-phosphate + UTP + H+ = UDP-N-acetyl- alpha-D-glucosamine + diphosphate
|
 |
 |
 |
 |
 |
 N-acetyl-alpha-D-glucosamine 1-phosphate
N-acetyl-alpha-D-glucosamine 1-phosphate
|
+
|
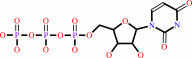 UTP
UTP
|
+
|
H(+)
|
=
|
UDP-N-acetyl- alpha-D-glucosamine
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
16:2657-2666
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Characterization of substrate binding and catalysis in the potential antibacterial target N-acetylglucosamine-1-phosphate uridyltransferase (GlmU).
|
|
I.Mochalkin,
S.Lightle,
Y.Zhu,
J.F.Ohren,
C.Spessard,
N.Y.Chirgadze,
C.Banotai,
M.Melnick,
L.McDowell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
N-Acetylglucosamine-1-phosphate uridyltransferase (GlmU) catalyzes the first
step in peptidoglycan biosynthesis in both Gram-positive and Gram-negative
bacteria. The products of the GlmU reaction are essential for bacterial
survival, making this enzyme an attractive target for antibiotic drug discovery.
A series of Haemophilus influenzae GlmU (hiGlmU) structures were determined by
X-ray crystallography in order to provide structural and functional insights
into GlmU activity and inhibition. The information derived from these structures
was combined with biochemical characterization of the K25A, Q76A, D105A, Y103A,
V223A, and E224A hiGlmU mutants in order to map these active-site residues to
catalytic activity of the enzyme and refine the mechanistic model of the GlmU
uridyltransferase reaction. These studies suggest that GlmU activity follows a
sequential substrate-binding order that begins with UTP binding noncovalently to
the GlmU enzyme. The uridyltransferase active site then remains in an open
apo-like conformation until N-acetylglucosamine-1-phosphate (GlcNAc-1-P) binds
and induces a conformational change at the GlcNAc-binding subsite. Following the
binding of GlcNAc-1-P to the UTP-charged uridyltransferase active site, the
non-esterified oxygen of GlcNAc-1-P performs a nucleophilic attack on the
alpha-phosphate group of UTP. The new data strongly suggest that the mechanism
of phosphotransfer in the uridyltransferase reaction in GlmU is primarily
through an associative mechanism with a pentavalent phosphate intermediate and
an inversion of stereochemistry. Finally, the structural and biochemical
characterization of the uridyltransferase active site and catalytic mechanism
described herein provides a basis for the structure-guided design of novel
antibacterial agents targeting GlmU activity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. hiGlmU uridyltransferase active site. (A) Stereoview of the (Fo-Fc) OMIT electron density maps of UDP-GlcNAc, UDP, and uridine bound to the
|
 |
Figure 4.
Figure 4. Structural insights into the mechanism of uridylation. (A) View of the GlmU uridyltransferase active site (open conformation) in the UDP-bound
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2007,
16,
2657-2666)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.F.Trempe,
S.Shenker,
G.Kozlov,
and
K.Gehring
(2011).
Self-association studies of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase from Escherichia coli.
|
| |
Protein Sci,
20,
745-752.
|
 |
|
|
|
|
 |
Z.Zhang,
E.M.Bulloch,
R.D.Bunker,
E.N.Baker,
and
C.J.Squire
(2009).
Structure and function of GlmU from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
275-283.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Mochalkin,
S.Lightle,
L.Narasimhan,
D.Bornemeier,
M.Melnick,
S.Vanderroest,
and
L.McDowell
(2008).
Structure of a small-molecule inhibitor complexed with GlmU from Haemophilus influenzae reveals an allosteric binding site.
|
| |
Protein Sci,
17,
577-582.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Yin,
C.R.Garen,
M.M.Cherney,
L.T.Cherney,
and
M.N.James
(2008).
Expression, purification and preliminary crystallographic analysis of N-acetylglucosamine-1-phosphate uridylyltransferase from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
805-808.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

