 |
PDBsum entry 1eu1

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1eu1
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.7.2.3
- trimethylamine-N-oxide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
trimethylamine + 2 Fe(III)-[cytochrome c] + H2O = trimethylamine N-oxide + 2 Fe(II)-[cytochrome c] + 3 H+
|
 |
 |
 |
 |
 |
 trimethylamine
trimethylamine
|
+
|
2
×
Fe(III)-[cytochrome c]
|
+
|
H2O
|
=
|
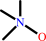 trimethylamine N-oxide
trimethylamine N-oxide
|
+
|
2
×
Fe(II)-[cytochrome c]
|
+
|
3
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mo-bis(molybdopterin guanine dinucleotide)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.8.5.3
- respiratory dimethylsulfoxide reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dimethyl sulfide + a menaquinone + H2O = dimethyl sulfoxide + a menaquinol
|
 |
 |
 |
 |
 |
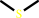 dimethyl sulfide
dimethyl sulfide
|
+
|
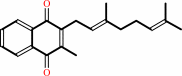 2
×
menaquinone
2
×
menaquinone
|
+
|
H2O
|
=
|
 dimethyl sulfoxide
dimethyl sulfoxide
|
+
|
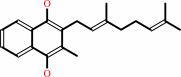 2
×
menaquinol
2
×
menaquinol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur; Mo-molybdopterin
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
Mo-molybdopterin
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
272:1615-1621
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination.
|
|
H.Schindelin,
C.Kisker,
J.Hilton,
K.V.Rajagopalan,
D.C.Rees.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The molybdoenzyme dimethylsulfoxide (DMSO) reductase contributes to the release
of dimethylsulfide, a compound that has been implicated in cloud nucleation and
global climate regulation. The crystal structure of DMSO reductase from
Rhodobacter sphaeroides reveals a monooxo molybdenum cofactor containing two
molybdopterin guanine dinucleotides that asymmetrically coordinate the
molybdenum through their dithiolene groups. One of the pterins exhibits
different coordination modes to the molybdenum between the oxidized and reduced
states, whereas the side chain oxygen of Ser147 coordinates the metal in both
states. The change in pterin coordination between the Mo(VI) and Mo(IV) forms
suggests a mechanism for substrate binding and reduction by this enzyme.
Sequence comparisons of DMSO reductase with a family of bacterial
oxotransferases containing molybdopterin guanine dinucleotide indicate a similar
polypeptide fold and active site with two molybdopterins within this family.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Mayilmurugan,
B.N.Harum,
M.Volpe,
A.F.Sax,
M.Palaniandavar,
and
N.C.Mösch-Zanetti
(2011).
Mechanistic insight into the reactivity of oxotransferases by novel asymmetric dioxomolybdenum(VI) model complexes.
|
| |
Chemistry,
17,
704-713.
|
 |
|
|
|
|
 |
C.A.Bayse
(2009).
Density-functional theory models of xanthine oxidoreductase activity: comparison of substrate tautomerization and protonation.
|
| |
Dalton Trans,
(),
2306-2314.
|
 |
|
|
|
|
 |
K.I.Chen,
A.G.McEwan,
and
P.V.Bernhardt
(2009).
Mediated electrochemistry of dimethyl sulfoxide reductase from Rhodobacter capsulatus.
|
| |
J Biol Inorg Chem,
14,
409-419.
|
 |
|
|
|
|
 |
M.J.Romão
(2009).
Molybdenum and tungsten enzymes: a crystallographic and mechanistic overview.
|
| |
Dalton Trans,
(),
4053-4068.
|
 |
|
|
|
|
 |
M.Neumann,
G.Mittelstädt,
C.Iobbi-Nivol,
M.Saggu,
F.Lendzian,
P.Hildebrandt,
and
S.Leimkühler
(2009).
A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli.
|
| |
FEBS J,
276,
2762-2774.
|
 |
|
|
|
|
 |
R.G.Hadt,
V.N.Nemykin,
J.G.Olsen,
and
P.Basu
(2009).
Comparative calculation of EPR spectral parameters in [Mo(V)OX4]-, [Mo(V)OX5]2-, and [Mo(V)OX4(H2O)]- complexes.
|
| |
Phys Chem Chem Phys,
11,
10377-10384.
|
 |
|
|
|
|
 |
U.Ryde,
C.Schulzke,
and
K.Starke
(2009).
Which functional groups of the molybdopterin ligand should be considered when modeling the active sites of the molybdenum and tungsten cofactors? A density functional theory study.
|
| |
J Biol Inorg Chem,
14,
1053-1064.
|
 |
|
|
|
|
 |
G.J.Workun,
K.Moquin,
R.A.Rothery,
and
J.H.Weiner
(2008).
Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold.
|
| |
Microbiol Mol Biol Rev,
72,
228.
|
 |
|
|
|
|
 |
H.Sugimoto,
and
H.Tsukube
(2008).
Chemical analogues relevant to molybdenum and tungsten enzyme reaction centres toward structural dynamics and reaction diversity.
|
| |
Chem Soc Rev,
37,
2609-2619.
|
 |
|
|
|
|
 |
M.Jormakka,
K.Yokoyama,
T.Yano,
M.Tamakoshi,
S.Akimoto,
T.Shimamura,
P.Curmi,
and
S.Iwata
(2008).
Molecular mechanism of energy conservation in polysulfide respiration.
|
| |
Nat Struct Mol Biol,
15,
730-737.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Fukuzumi,
and
T.Kojima
(2008).
Control of redox reactivity of flavin and pterin coenzymes by metal ion coordination and hydrogen bonding.
|
| |
J Biol Inorg Chem,
13,
321-333.
|
 |
|
|
|
|
 |
T.Reda,
C.M.Plugge,
N.J.Abram,
and
J.Hirst
(2008).
Reversible interconversion of carbon dioxide and formate by an electroactive enzyme.
|
| |
Proc Natl Acad Sci U S A,
105,
10654-10658.
|
 |
|
|
|
|
 |
Y.Qiu,
R.Zhang,
T.A.Binkowski,
V.Tereshko,
A.Joachimiak,
and
A.Kossiakoff
(2008).
The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
|
| |
Proteins,
71,
525-533.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Zhang,
and
V.N.Gladyshev
(2008).
Molybdoproteomes and evolution of molybdenum utilization.
|
| |
J Mol Biol,
379,
881-899.
|
 |
|
|
|
|
 |
E.V.Morozkina,
and
R.A.Zvyagilskaya
(2007).
Nitrate reductases: structure, functions, and effect of stress factors.
|
| |
Biochemistry (Mosc),
72,
1151-1160.
|
 |
|
|
|
|
 |
G.B.Seiffert,
G.M.Ullmann,
A.Messerschmidt,
B.Schink,
P.M.Kroneck,
and
O.Einsle
(2007).
Structure of the non-redox-active tungsten/[4Fe:4S] enzyme acetylene hydratase.
|
| |
Proc Natl Acad Sci U S A,
104,
3073-3077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Lyashenko,
G.Saischek,
A.Pal,
R.Herbst-Irmer,
and
N.C.Mösch-Zanetti
(2007).
Molecular oxygen activation by a molybdenum(IV) monooxo bis(beta-ketiminato) complex.
|
| |
Chem Commun (Camb),
(),
701-703.
|
 |
|
|
|
|
 |
G.N.George,
K.J.Nelson,
H.H.Harris,
C.J.Doonan,
and
K.V.Rajagopalan
(2007).
Interaction of product analogues with the active site of rhodobacter sphaeroides dimethyl sulfoxide reductase.
|
| |
Inorg Chem,
46,
3097-3104.
|
 |
|
|
|
|
 |
M.Hofmann
(2007).
Density functional theory studies of model complexes for molybdenum-dependent nitrate reductase active sites.
|
| |
J Biol Inorg Chem,
12,
989.
|
 |
|
|
|
|
 |
X.Ma,
C.Schulzke,
H.G.Schmidt,
and
M.Noltemeyer
(2007).
Structural, electrochemical and oxygen atom transfer properties of a molybdenum selenoether complex [Mo2O4(OC3H6SeC3H6O)2] and its thioether analogue [Mo2O4(OC3H6SC3H6O)2].
|
| |
Dalton Trans,
(),
1773-1780.
|
 |
|
|
|
|
 |
A.Thapper,
A.Behrens,
J.Fryxelius,
M.H.Johansson,
F.Prestopino,
M.Czaun,
D.Rehder,
and
E.Nordlander
(2005).
Synthesis and characterization of molybdenum oxo complexes of two tripodal ligands: reactivity studies of a functional model for molybdenum oxotransferases.
|
| |
Dalton Trans,
(),
3566-3571.
|
 |
|
|
|
|
 |
J.A.Müller,
and
S.DasSarma
(2005).
Genomic analysis of anaerobic respiration in the archaeon Halobacterium sp. strain NRC-1: dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors.
|
| |
J Bacteriol,
187,
1659-1667.
|
 |
|
|
|
|
 |
A.Messerschmidt,
H.Niessen,
D.Abt,
O.Einsle,
B.Schink,
and
P.M.Kroneck
(2004).
Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols.
|
| |
Proc Natl Acad Sci U S A,
101,
11571-11576.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.R.Luckarift,
H.Dalton,
N.D.Sharma,
D.R.Boyd,
and
R.A.Holt
(2004).
Isolation and characterisation of bacterial strains containing enantioselective DMSO reductase activity: application to the kinetic resolution of racemic sulfoxides.
|
| |
Appl Microbiol Biotechnol,
65,
678-685.
|
 |
|
|
|
|
 |
J.J.Moura,
C.D.Brondino,
J.Trincão,
and
M.J.Romão
(2004).
Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases.
|
| |
J Biol Inorg Chem,
9,
791-799.
|
 |
|
|
|
|
 |
J.Kuper,
A.Llamas,
H.J.Hecht,
R.R.Mendel,
and
G.Schwarz
(2004).
Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism.
|
| |
Nature,
430,
803-806.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Tomiki,
and
N.Saitou
(2004).
Phylogenetic analysis of proteins associated in the four major energy metabolism systems: photosynthesis, aerobic respiration, denitrification, and sulfur respiration.
|
| |
J Mol Evol,
59,
158-176.
|
 |
|
|
|
|
 |
M.G.Bertero,
R.A.Rothery,
M.Palak,
C.Hou,
D.Lim,
F.Blasco,
J.H.Weiner,
and
N.C.Strynadka
(2003).
Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A.
|
| |
Nat Struct Biol,
10,
681-687.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Jormakka,
B.Byrne,
and
S.Iwata
(2003).
Formate dehydrogenase--a versatile enzyme in changing environments.
|
| |
Curr Opin Struct Biol,
13,
418-423.
|
 |
|
|
|
|
 |
C.A.McDevitt,
P.Hugenholtz,
G.R.Hanson,
and
A.G.McEwan
(2002).
Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes.
|
| |
Mol Microbiol,
44,
1575-1587.
|
 |
|
|
|
|
 |
C.Sandu,
and
R.Brandsch
(2002).
Functional analysis of the Escherichia coli molybdopterin cofactor biosynthesis protein MoeA by site-directed mutagenesis.
|
| |
Biol Chem,
383,
319-323.
|
 |
|
|
|
|
 |
J.Simon
(2002).
Enzymology and bioenergetics of respiratory nitrite ammonification.
|
| |
FEMS Microbiol Rev,
26,
285-309.
|
 |
|
|
|
|
 |
R.Hille
(2002).
Molybdenum and tungsten in biology.
|
| |
Trends Biochem Sci,
27,
360-367.
|
 |
|
|
|
|
 |
A.F.Bell,
X.He,
J.P.Ridge,
G.R.Hanson,
A.G.McEwan,
and
P.J.Tonge
(2001).
Active site heterogeneity in dimethyl sulfoxide reductase from Rhodobacter capsulatus revealed by Raman spectroscopy.
|
| |
Biochemistry,
40,
440-448.
|
 |
|
|
|
|
 |
K.Heffron,
C.Léger,
R.A.Rothery,
J.H.Weiner,
and
F.A.Armstrong
(2001).
Determination of an optimal potential window for catalysis by E. coli dimethyl sulfoxide reductase and hypothesis on the role of Mo(V) in the reaction pathway.
|
| |
Biochemistry,
40,
3117-3126.
|
 |
|
|
|
|
 |
V.V.Pollock,
and
M.J.Barber
(2001).
Kinetic and mechanistic properties of biotin sulfoxide reductase.
|
| |
Biochemistry,
40,
1430-1440.
|
 |
|
|
|
|
 |
C.A.Temple,
G.N.George,
J.C.Hilton,
M.J.George,
R.C.Prince,
M.J.Barber,
and
K.V.Rajagopalan
(2000).
Structure of the molybdenum site of Rhodobacter sphaeroides biotin sulfoxide reductase.
|
| |
Biochemistry,
39,
4046-4052.
|
 |
|
|
|
|
 |
R.C.Bray,
B.Adams,
A.T.Smith,
B.Bennett,
and
S.Bailey
(2000).
Reversible dissociation of thiolate ligands from molybdenum in an enzyme of the dimethyl sulfoxide reductase family.
|
| |
Biochemistry,
39,
11258-11269.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.T.Lowther,
N.Brot,
H.Weissbach,
and
B.W.Matthews
(2000).
Structure and mechanism of peptide methionine sulfoxide reductase, an "anti-oxidation" enzyme.
|
| |
Biochemistry,
39,
13307-13312.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.T.Lowther,
N.Brot,
H.Weissbach,
J.F.Honek,
and
B.W.Matthews
(2000).
Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase.
|
| |
Proc Natl Acad Sci U S A,
97,
6463-6468.
|
 |
|
|
|
|
 |
B.Adams,
A.T.Smith,
S.Bailey,
A.G.McEwan,
and
R.C.Bray
(1999).
Reactions of dimethylsulfoxide reductase from Rhodobacter capsulatus with dimethyl sulfide and with dimethyl sulfoxide: complexities revealed by conventional and stopped-flow spectrophotometry.
|
| |
Biochemistry,
38,
8501-8511.
|
 |
|
|
|
|
 |
C.S.Butler,
J.M.Charnock,
B.Bennett,
H.J.Sears,
A.J.Reilly,
S.J.Ferguson,
C.D.Garner,
D.J.Lowe,
A.J.Thomson,
B.C.Berks,
and
D.J.Richardson
(1999).
Models for molybdenum coordination during the catalytic cycle of periplasmic nitrate reductase from Paracoccus denitrificans derived from EPR and EXAFS spectroscopy.
|
| |
Biochemistry,
38,
9000-9012.
|
 |
|
|
|
|
 |
D.Baas,
and
J.Rétey
(1999).
Cloning, sequencing and heterologous expression of pyrogallol-phloroglucinol transhydroxylase from Pelobacter acidigallici.
|
| |
Eur J Biochem,
265,
896-901.
|
 |
|
|
|
|
 |
J.Buc,
C.L.Santini,
R.Giordani,
M.Czjzek,
L.F.Wu,
and
G.Giordano
(1999).
Enzymatic and physiological properties of the tungsten-substituted molybdenum TMAO reductase from Escherichia coli.
|
| |
Mol Microbiol,
32,
159-168.
|
 |
|
|
|
|
 |
M.Hensel,
A.P.Hinsley,
T.Nikolaus,
G.Sawers,
and
B.C.Berks
(1999).
The genetic basis of tetrathionate respiration in Salmonella typhimurium.
|
| |
Mol Microbiol,
32,
275-287.
|
 |
|
|
|
|
 |
M.J.Almendra,
C.D.Brondino,
O.Gavel,
A.S.Pereira,
P.Tavares,
S.Bursakov,
R.Duarte,
J.Caldeira,
J.J.Moura,
and
I.Moura
(1999).
Purification and characterization of a tungsten-containing formate dehydrogenase from Desulfovibrio gigas.
|
| |
Biochemistry,
38,
16366-16372.
|
 |
|
|
|
|
 |
R.U.Meckenstock,
R.Krieger,
S.Ensign,
P.M.Kroneck,
and
B.Schink
(1999).
Acetylene hydratase of Pelobacter acetylenicus. Molecular and spectroscopic properties of the tungsten iron-sulfur enzyme.
|
| |
Eur J Biochem,
264,
176-182.
|
 |
|
|
|
|
 |
A.Hasona,
R.M.Ray,
and
K.T.Shanmugam
(1998).
Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli.
|
| |
J Bacteriol,
180,
1466-1472.
|
 |
|
|
|
|
 |
A.Magalon,
M.Asso,
B.Guigliarelli,
R.A.Rothery,
P.Bertrand,
G.Giordano,
and
F.Blasco
(1998).
Molybdenum cofactor properties and [Fe-S] cluster coordination in Escherichia coli nitrate reductase A: investigation by site-directed mutagenesis of the conserved his-50 residue in the NarG subunit.
|
| |
Biochemistry,
37,
7363-7370.
|
 |
|
|
|
|
 |
C.Kisker,
H.Schindelin,
D.Baas,
J.Rétey,
R.U.Meckenstock,
and
P.M.Kroneck
(1998).
A structural comparison of molybdenum cofactor-containing enzymes.
|
| |
FEMS Microbiol Rev,
22,
503-521.
|
 |
|
|
|
|
 |
C.L.Santini,
B.Ize,
A.Chanal,
M.Müller,
G.Giordano,
and
L.F.Wu
(1998).
A novel sec-independent periplasmic protein translocation pathway in Escherichia coli.
|
| |
EMBO J,
17,
101-112.
|
 |
|
|
|
|
 |
D.M.Luykx,
J.A.Duine,
and
S.de Vries
(1998).
Molybdopterin radical in bacterial aldehyde dehydrogenases.
|
| |
Biochemistry,
37,
11366-11375.
|
 |
|
|
|
|
 |
J.McMaster,
and
J.H.Enemark
(1998).
The active sites of molybdenum- and tungsten-containing enzymes.
|
| |
Curr Opin Chem Biol,
2,
201-207.
|
 |
|
|
|
|
 |
M.Matsuzaki,
Y.Kiso,
I.Yamamoto,
and
T.Satoh
(1998).
Isolation of a periplasmic molecular chaperone-like protein of Rhodobacter sphaeroides f. sp. denitrificans that is homologous to the dipeptide transport protein DppA of Escherichia coli.
|
| |
J Bacteriol,
180,
2718-2722.
|
 |
|
|
|
|
 |
N.J.Mouncey,
and
S.Kaplan
(1998).
Redox-dependent gene regulation in Rhodobacter sphaeroides 2.4.1(T): effects on dimethyl sulfoxide reductase (dor) gene expression.
|
| |
J Bacteriol,
180,
5612-5618.
|
 |
|
|
|
|
 |
R.Hille,
J.Rétey,
U.Bartlewski-Hof,
and
W.Reichenbecher
(1998).
Mechanistic aspects of molybdenum-containing enzymes.
|
| |
FEMS Microbiol Rev,
22,
489-501.
|
 |
|
|
|
|
 |
R.W.Frazee,
A.M.Orville,
K.B.Dolbeare,
H.Yu,
D.H.Ohlendorf,
and
J.D.Lipscomb
(1998).
The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
|
| |
Biochemistry,
37,
2131-2144.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.V.Khangulov,
V.N.Gladyshev,
G.C.Dismukes,
and
T.C.Stadtman
(1998).
Selenium-containing formate dehydrogenase H from Escherichia coli: a molybdopterin enzyme that catalyzes formate oxidation without oxygen transfer.
|
| |
Biochemistry,
37,
3518-3528.
|
 |
|
|
|
|
 |
T.Palmer,
I.P.Goodfellow,
R.E.Sockett,
A.G.McEwan,
and
D.H.Boxer
(1998).
Characterisation of the mob locus from Rhodobacter sphaeroides required for molybdenum cofactor biosynthesis.
|
| |
Biochim Biophys Acta,
1395,
135-140.
|
 |
|
|
|
|
 |
U.Ermler,
W.Grabarse,
S.Shima,
M.Goubeaud,
and
R.K.Thauer
(1998).
Active sites of transition-metal enzymes with a focus on nickel.
|
| |
Curr Opin Struct Biol,
8,
749-758.
|
 |
|
|
|
|
 |
C.Kisker,
H.Schindelin,
A.Pacheco,
W.A.Wehbi,
R.M.Garrett,
K.V.Rajagopalan,
J.H.Enemark,
and
D.C.Rees
(1997).
Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase.
|
| |
Cell,
91,
973-983.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Kisker,
H.Schindelin,
and
D.C.Rees
(1997).
Molybdenum-cofactor-containing enzymes: structure and mechanism.
|
| |
Annu Rev Biochem,
66,
233-267.
|
 |
|
|
|
|
 |
J.C.Boyington,
V.N.Gladyshev,
S.V.Khangulov,
T.C.Stadtman,
and
P.D.Sun
(1997).
Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
|
| |
Science,
275,
1305-1308.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.S.Solomon,
I.Lane,
G.R.Hanson,
and
A.G.McEwan
(1997).
Characterisation of the pterin molybdenum cofactor in dimethylsulfoxide reductase of Rhodobacter capsulatus.
|
| |
Eur J Biochem,
246,
200-203.
|
 |
|
|
|
|
 |
W.G.Zumft
(1997).
Cell biology and molecular basis of denitrification.
|
| |
Microbiol Mol Biol Rev,
61,
533-616.
|
 |
|
|
|
|
 |
A.Hochheimer,
D.Linder,
R.K.Thauer,
and
R.Hedderich
(1996).
The molybdenum formylmethanofuran dehydrogenase operon and the tungsten formylmethanofuran dehydrogenase operon from Methanobacterium thermoautotrophicum. Structures and transcriptional regulation.
|
| |
Eur J Biochem,
242,
156-162.
|
 |
|
|
|
|
 |
A.L.Shaw,
G.R.Hanson,
and
A.G.McEwan
(1996).
Cloning and sequence analysis of the dimethylsulfoxide reductase structural gene from Rhodobacter capsulatus.
|
| |
Biochim Biophys Acta,
1276,
176-180.
|
 |
|
|
|
|
 |
A.Volbeda,
J.C.Fontecilla-Camps,
and
M.Frey
(1996).
Novel metal sites in protein structures.
|
| |
Curr Opin Struct Biol,
6,
804-812.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

