
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 0 more)
875 a.a.
(+ 0 more)
875 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 0 more)
274 a.a.
(+ 0 more)
274 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
 |
Obsolete entry |
 |
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of pyrogallol-phloroglucinol transhydroxylase from pelobacter acidigallici
|
|
Structure:
|
 |
Pyrogallol hydroxytransferase large subunit. Chain: a, c, e, g, i, k. Synonym: transhydroxylase alpha subunit. Pyrogallol hydroxytransferase small subunit. Chain: b, d, f, h, j, l. Synonym: transhydroxylase beta subunit. Ec: 1.97.1.2
|
|
Source:
|
 |
Pelobacter acidigallici. Organism_taxid: 35816. Strain: magal 2(dsm 2377). Strain: magal 2(dsm 2377)
|
|
Biol. unit:
|
 |
Dodecamer (from
)
|
|
Resolution:
|
 |
|
2.35Å
|
R-factor:
|
0.198
|
R-free:
|
0.254
|
|
|
Authors:
|
 |
A.Messerschmidt,H.Niessen,D.Abt,O.Einsle,B.Schink,P.M.H.Kroneck
|
Key ref:

|
 |
A.Messerschmidt
et al.
(2004).
Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols.
Proc Natl Acad Sci U S A,
101,
11571-11576.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Jun-04
|
Release date:
|
10-Aug-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, C, E, G, I, K:
E.C.1.97.1.2
- pyrogallol hydroxytransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1,2,3,5-tetrahydroxybenzene + 1,2,3-trihydroxybenzene = 1,2,3,5- tetrahydroxybenzene + 1,3,5-trihydroxybenzene
|
 |
 |
 |
 |
 |
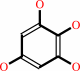 1,2,3,5-tetrahydroxybenzene
1,2,3,5-tetrahydroxybenzene
|
+
|
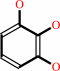 1,2,3-trihydroxybenzene
1,2,3-trihydroxybenzene
|
=
|
 1,2,3,5- tetrahydroxybenzene
1,2,3,5- tetrahydroxybenzene
|
+
|
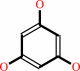 1,3,5-trihydroxybenzene
1,3,5-trihydroxybenzene
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mo cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
101:11571-11576
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols.
|
|
A.Messerschmidt,
H.Niessen,
D.Abt,
O.Einsle,
B.Schink,
P.M.Kroneck.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The Mo enzyme transhydroxylase from the anaerobic microorganism Pelobacter
acidigallici catalyzes the conversion of pyrogallol to phloroglucinol. Such
trihydroxybenzenes and their derivatives represent important building blocks of
plant polymers. None of the transferred hydroxyl groups originates from water
during transhydroxylation; instead a cosubstrate, such as
1,2,3,5-tetrahydroxybenzene, is used in a reaction without apparent electron
transfer. Here, we report on the crystal structure of the enzyme in the reduced
Mo(IV) state, which we solved by single anomalous-diffraction technique. It
represents the largest structure (1,149 amino acid residues per molecule, 12
independent molecules per unit cell), which has been solved so far by single
anomalous-diffraction technique. Tranhydroxylase is a heterodimer, with the
active Mo-molybdopterin guanine dinucleotide (MGD)(2) site in the alpha-subunit,
and three [4Fe-4S] centers in the beta-subunit. The latter subunit carries a
seven-stranded, mainly antiparallel beta-barrel domain. We propose a scheme for
the transhydroxylation reaction based on 3D structures of complexes of the
enzyme with various polyphenols serving either as substrate or inhibitor.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Overall structure of TH. The  -subunit domains I-IV
are shown in magenta, blue, red, and cream, respectively. The -subunit domains I-IV
are shown in magenta, blue, red, and cream, respectively. The
 -subunit domains I and
II are shown in orange and pink, respectively, and domain III is
shown in green. The Mo and MGD cofactors are shown as
ball-and-stick models, and the three [4Fe--4S] clusters are
shown as red (Fe) and yellow (S) spheres. The figure was made
with BOBSCRIPT (31) and RASTER3D (32). -subunit domains I and
II are shown in orange and pink, respectively, and domain III is
shown in green. The Mo and MGD cofactors are shown as
ball-and-stick models, and the three [4Fe--4S] clusters are
shown as red (Fe) and yellow (S) spheres. The figure was made
with BOBSCRIPT (31) and RASTER3D (32).
|
 |
Figure 2.
Fig. 2. Solid-surface-electrostatic potential
representation of TH displaying the access channel for substrate
and cosubstrate. The electrostatic surface potentials are
contoured from -10 (red) to 10 K[B]T/e (blue). The figure was
made with GRASP (34) and RASTER3D (32).
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Carmona,
M.T.Zamarro,
B.Blázquez,
G.Durante-Rodríguez,
J.F.Juárez,
J.A.Valderrama,
M.J.Barragán,
J.L.García,
and
E.Díaz
(2009).
Anaerobic catabolism of aromatic compounds: a genetic and genomic view.
|
| |
Microbiol Mol Biol Rev,
73,
71.
|
 |
|
|
|
|
 |
H.Sugimoto,
and
H.Tsukube
(2008).
Chemical analogues relevant to molybdenum and tungsten enzyme reaction centres toward structural dynamics and reaction diversity.
|
| |
Chem Soc Rev,
37,
2609-2619.
|
 |
|
|
|
|
 |
G.B.Seiffert,
G.M.Ullmann,
A.Messerschmidt,
B.Schink,
P.M.Kroneck,
and
O.Einsle
(2007).
Structure of the non-redox-active tungsten/[4Fe:4S] enzyme acetylene hydratase.
|
| |
Proc Natl Acad Sci U S A,
104,
3073-3077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.I.Darley,
J.A.Hellstern,
J.I.Medina-Bellver,
S.Marqués,
B.Schink,
and
B.Philipp
(2007).
Heterologous expression and identification of the genes involved in anaerobic degradation of 1,3-dihydroxybenzene (resorcinol) in Azoarcus anaerobius.
|
| |
J Bacteriol,
189,
3824-3833.
|
 |
|
|
|
|
 |
D.P.Kloer,
C.Hagel,
J.Heider,
and
G.E.Schulz
(2006).
Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum.
|
| |
Structure,
14,
1377-1388.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Boll,
B.Schink,
A.Messerschmidt,
and
P.M.Kroneck
(2005).
Novel bacterial molybdenum and tungsten enzymes: three-dimensional structure, spectroscopy, and reaction mechanism.
|
| |
Biol Chem,
386,
999.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

