 |
PDBsum entry 1d2f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.4.1.13
- cysteine-S-conjugate beta-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an S-substituted L-cysteine + H2O = a thiol + pyruvate + NH4+
|
 |
 |
 |
 |
 |
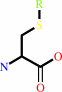 S-substituted L-cysteine
S-substituted L-cysteine
|
+
|
H2O
|
=
|
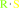 thiol
thiol
|
+
|
 pyruvate
pyruvate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
19:831-842
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray structure of MalY from Escherichia coli: a pyridoxal 5'-phosphate-dependent enzyme acting as a modulator in mal gene expression.
|
|
T.Clausen,
A.Schlegel,
R.Peist,
E.Schneider,
C.Steegborn,
Y.S.Chang,
A.Haase,
G.P.Bourenkov,
H.D.Bartunik,
W.Boos.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
MalY represents a bifunctional pyridoxal 5'-phosphate-dependent enzyme acting as
a beta-cystathionase and as a repressor of the maltose regulon. Here we present
the crystal structures of wild-type and A221V mutant protein. Each subunit of
the MalY dimer is composed of a large pyridoxal 5'-phosphate-binding domain and
a small domain similar to aminotransferases. The structural alignment with
related enzymes identifies residues that are generally responsible for
beta-lyase activity and depicts a unique binding mode of the pyridoxal
5'-phosphate correlated with a larger, more flexible substrate-binding pocket.
In a screen for MalY mutants with reduced mal repressor properties, mutations
occurred in three clusters: I, 83-84; II, 181-189 and III, 215-221, which
constitute a clearly distinguished region in the MalY crystal structure far away
from the cofactor. The tertiary structure of one of these mutants (A221V)
demonstrates that positional rearrangements are indeed restricted to regions I,
II and III. Therefore, we propose that a direct protein-protein interaction with
MalT, the central transcriptional activator of the maltose system, underlies
MalY-dependent repression of the maltose system.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4 MalT-binding site. (A) The active dimer of MalY
illustrating the location of the MalT interaction patch. The C^
 traces
of both monomers (white and green) are overlaid with a
transparent surface. The MalT interaction regions are emphasized
by a solid surface that was defined on the basis of the negative
repressor mutants (drawn in red). The PLP cofactor is shown in a
van der Waals representation. Note that the MalT-binding surface
and the active site entrance to the PLP cofactor are located on
opposite sides of the individual MalY monomers. (B) Spatial
structure of the MalT-binding patch, which is constructed from
the three segments I, II and III as described in the text. The C
atoms of segments I, II and III are coloured orange (residues
81–85), white (residues 179–191) and green (residues
212–222), respectively. For each segment, the most important
residue regarding MalT repression (Table II) is labelled. The
model is overlaid with a transparent surface that is colour
coded by atom type. (C) Overlay of the wild-type and A221V MalT
interaction segments I, II and III. The wild-type model and the
corresponding surface are in white, the A221V mutant in green.
Obviously, the mutation Ala221 to Val221 results in a concerted
structural reorientation of all three segments. The orientations
of (B) and (C) are identical. traces
of both monomers (white and green) are overlaid with a
transparent surface. The MalT interaction regions are emphasized
by a solid surface that was defined on the basis of the negative
repressor mutants (drawn in red). The PLP cofactor is shown in a
van der Waals representation. Note that the MalT-binding surface
and the active site entrance to the PLP cofactor are located on
opposite sides of the individual MalY monomers. (B) Spatial
structure of the MalT-binding patch, which is constructed from
the three segments I, II and III as described in the text. The C
atoms of segments I, II and III are coloured orange (residues
81–85), white (residues 179–191) and green (residues
212–222), respectively. For each segment, the most important
residue regarding MalT repression (Table II) is labelled. The
model is overlaid with a transparent surface that is colour
coded by atom type. (C) Overlay of the wild-type and A221V MalT
interaction segments I, II and III. The wild-type model and the
corresponding surface are in white, the A221V mutant in green.
Obviously, the mutation Ala221 to Val221 results in a concerted
structural reorientation of all three segments. The orientations
of (B) and (C) are identical.
|
 |
Figure 5.
Figure 5 Active site entrances of MalY (left) and CBL (right).
The orientation and scaling of both figures are identical. The
corresponding PLP cofactors are shown in a van der Waals
representation below the surface. Part of the phosphate group of
the MalY cofactor is directly accessible in the active site
cleft.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2000,
19,
831-842)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Lengsfeld,
S.Schönert,
R.Dippel,
and
W.Boos
(2009).
Glucose- and glucokinase-controlled mal gene expression in Escherichia coli.
|
| |
J Bacteriol,
191,
701-712.
|
 |
|
|
|
|
 |
C.Tanous,
O.Soutourina,
B.Raynal,
M.F.Hullo,
P.Mervelet,
A.M.Gilles,
P.Noirot,
A.Danchin,
P.England,
and
I.Martin-Verstraete
(2008).
The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis.
|
| |
J Biol Chem,
283,
35551-35560.
|
 |
|
|
|
|
 |
Y.Yoshida,
S.Ito,
T.Sasaki,
M.Kishi,
M.Kurota,
A.Suwabe,
K.Kunimatsu,
and
H.Kato
(2008).
Molecular and enzymatic characterization of betaC-S lyase in Streptococcus constellatus.
|
| |
Oral Microbiol Immunol,
23,
245-253.
|
 |
|
|
|
|
 |
S.Lima,
R.Khristoforov,
C.Momany,
and
R.S.Phillips
(2007).
Crystal structure of Homo sapiens kynureninase.
|
| |
Biochemistry,
46,
2735-2744.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Awano,
M.Wada,
H.Mori,
S.Nakamori,
and
H.Takagi
(2005).
Identification and functional analysis of Escherichia coli cysteine desulfhydrases.
|
| |
Appl Environ Microbiol,
71,
4149-4152.
|
 |
|
|
|
|
 |
A.Paiardini,
F.Bossa,
and
S.Pascarella
(2004).
Evolutionarily conserved regions and hydrophobic contacts at the superfamily level: The case of the fold-type I, pyridoxal-5'-phosphate-dependent enzymes.
|
| |
Protein Sci,
13,
2992-3005.
|
 |
|
|
|
|
 |
N.Joly,
A.Böhm,
W.Boos,
and
E.Richet
(2004).
MalK, the ATP-binding cassette component of the Escherichia coli maltodextrin transporter, inhibits the transcriptional activator malt by antagonizing inducer binding.
|
| |
J Biol Chem,
279,
33123-33130.
|
 |
|
|
|
|
 |
N.Sorrentino,
G.De Simone,
V.Menchise,
L.Mandrich,
M.Rossi,
G.Manco,
and
C.Pedone
(2003).
Crystallization and preliminary X-ray diffraction studies of Aes acetyl-esterase from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1846-1848.
|
 |
|
|
|
|
 |
R.Percudani,
and
A.Peracchi
(2003).
A genomic overview of pyridoxal-phosphate-dependent enzymes.
|
| |
EMBO Rep,
4,
850-854.
|
 |
|
|
|
|
 |
A.Schlegel,
O.Danot,
E.Richet,
T.Ferenci,
and
W.Boos
(2002).
The N terminus of the Escherichia coli transcription activator MalT is the domain of interaction with MalY.
|
| |
J Bacteriol,
184,
3069-3077.
|
 |
|
|
|
|
 |
L.Mandrich,
E.Caputo,
B.M.Martin,
M.Rossi,
and
G.Manco
(2002).
The Aes protein and the monomeric alpha-galactosidase from Escherichia coli form a non-covalent complex. Implications for the regulation of carbohydrate metabolism.
|
| |
J Biol Chem,
277,
48241-48247.
|
 |
|
|
|
|
 |
M.Bertoldi,
B.Cellini,
T.Clausen,
and
C.B.Voltattorni
(2002).
Spectroscopic and kinetic analyses reveal the pyridoxal 5'-phosphate binding mode and the catalytic features of Treponema denticola cystalysin.
|
| |
Biochemistry,
41,
9153-9164.
|
 |
|
|
|
|
 |
S.Le Bouder-Langevin,
I.Capron-Montaland,
R.De Rosa,
and
B.Labedan
(2002).
A strategy to retrieve the whole set of protein modules in microbial proteomes.
|
| |
Genome Res,
12,
1961-1973.
|
 |
|
|
|
|
 |
K.Haruyama,
T.Nakai,
I.Miyahara,
K.Hirotsu,
H.Mizuguchi,
H.Hayashi,
and
H.Kagamiyama
(2001).
Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
|
| |
Biochemistry,
40,
4633-4644.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.M.Wick,
M.Quadroni,
and
T.Egli
(2001).
Short- and long-term changes in proteome composition and kinetic properties in a culture of Escherichia coli during transition from glucose-excess to glucose-limited growth conditions in continuous culture and vice versa.
|
| |
Environ Microbiol,
3,
588-599.
|
 |
|
|
|
|
 |
H.I.Krupka,
R.Huber,
S.C.Holt,
and
T.Clausen
(2000).
Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
|
| |
EMBO J,
19,
3168-3178.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Feng,
M.K.Geck,
A.C.Eliot,
and
J.F.Kirsch
(2000).
Aminotransferase activity and bioinformatic analysis of 1-aminocyclopropane-1-carboxylate synthase.
|
| |
Biochemistry,
39,
15242-15249.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

