 |
PDBsum entry 1akc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase(aminotransferase)
|
PDB id
|
|

|

|
1akc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.1
- aspartate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 oxaloacetate
oxaloacetate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PPE)
matches with 57.69% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.7
- kynurenine--oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-kynurenine + 2-oxoglutarate = kynurenate + L-glutamate + H2O
|
 |
 |
 |
 |
 |
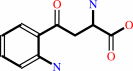 L-kynurenine
L-kynurenine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
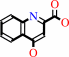 kynurenate
kynurenate
|
+
|
 L-glutamate
L-glutamate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PPE)
matches with 57.69% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
34:405-414
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the catalytic activity of aspartate aminotransferase K258H lacking the pyridoxal 5'-phosphate-binding lysine residue.
|
|
V.N.Malashkevich,
J.Jäger,
M.Ziak,
U.Sauder,
H.Gehring,
P.Christen,
J.N.Jansonius.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Chicken mitochondrial and Escherichia coli aspartate aminotransferases K258H, in
which the active site lysine residue has been exchanged for a histidine residue,
retain partial catalytic competence [Ziak et al. (1993) Eur. J. Biochem. 211,
475-484]. Mutant PLP and PMP holoenzymes and the complexes of the latter (E.
coli enzyme) with sulfate and 2-oxoglutarate, as well as complexes of the
mitochondrial apoenzyme with N-(5'-phosphopyridoxyl)-L-aspartate or
N-(5'-phosphopyridoxyl)-L-glutamate, were crystallized and analyzed by means of
X-ray crystallography in order to examine how the side chain of histidine 258
can substitute as a general acid/base catalyst of the aldimine-ketimine
tautomerization in enzymic transamination. The structures have been solved and
refined at resolutions between 2.1 and 2.8 A. Both the closed and the open
conformations, identical to those of the wild-type enzyme, were observed,
indicating that the mutant enzymes of both species exhibit the same
conformational flexibility as the wild-type enzymes, although in AspAT K258H the
equilibrium is somewhat shifted toward the open conformation. The replacement of
the active site K258 by a histidine residue resulted only in local structural
adaptations necessary to accommodate the imidazole ring. The catalytic
competence of the mutant enzyme, which in the forward half-reaction is 0.1% of
that of the wild-type enzyme, suggests that the imidazole group is involved in
the aldimine-ketimine tautomerization. However, the imidazole ring of H258 is
too far away from C alpha and C4' of the coenzyme-substrate adduct for direct
proton transfer, suggesting that the 1,3-prototropic shift is mediated by a
water molecule. Although there is enough space for a water molecule in this
area, it has not been detected. Dynamic fluctuations of the protein matrix might
transiently open a channel, giving a water molecule fleeting access to the
active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.J.Liao,
K.H.Chin,
C.H.Lin,
P.S.Tsai,
P.C.Lyu,
C.C.Young,
A.H.Wang,
and
S.H.Chou
(2008).
Crystal structure of DFA0005 complexed with alpha-ketoglutarate: a novel member of the ICL/PEPM superfamily from alkali-tolerant Deinococcus ficus.
|
| |
Proteins,
73,
362-371.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Golinelli-Pimpaneau,
C.Lüthi,
and
P.Christen
(2006).
Structural basis for D-amino acid transamination by the pyridoxal 5'-phosphate-dependent catalytic antibody 15A9.
|
| |
J Biol Chem,
281,
23969-23977.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.K.Hsu,
H.H.Lo,
C.H.Kao,
D.S.Lee,
and
W.H.Hsu
(2006).
Enantioselective synthesis of L-homophenylalanine by whole cells of recombinant Escherichia coli expressing L-aminoacylase and N-acylamino acid racemase genes from Deinococcus radiodurans BCRC12827.
|
| |
Biotechnol Prog,
22,
1578-1584.
|
 |
|
|
|
|
 |
H.H.Lo,
S.K.Hsu,
W.D.Lin,
N.L.Chan,
and
W.H.Hsu
(2005).
Asymmetrical synthesis of L-homophenylalanine using engineered Escherichia coli aspartate aminotransferase.
|
| |
Biotechnol Prog,
21,
411-415.
|
 |
|
|
|
|
 |
B.Cellini,
M.Bertoldi,
A.Paiardini,
S.D'Aguanno,
and
C.B.Voltattorni
(2004).
Site-directed mutagenesis provides insight into racemization and transamination of alanine catalyzed by Treponema denticola cystalysin.
|
| |
J Biol Chem,
279,
36898-36905.
|
 |
|
|
|
|
 |
J.Singh,
G.A.Khan,
L.Kinarsky,
H.Cheng,
J.Wilken,
K.H.Choi,
E.Bedows,
S.Sherman,
and
P.W.Cheng
(2004).
Identification of disulfide bonds among the nine core 2 N-acetylglucosaminyltransferase-M cysteines conserved in the mucin beta6-N-acetylglucosaminyltransferase family.
|
| |
J Biol Chem,
279,
38969-38977.
|
 |
|
|
|
|
 |
A.Poupon,
F.Jebai,
G.Labesse,
F.Gros,
J.Thibault,
J.P.Mornon,
and
M.Krieger
(1999).
Structure modelling and site-directed mutagenesis of the rat aromatic L-amino acid pyridoxal 5'-phosphate-dependent decarboxylase: a functional study.
|
| |
Proteins,
37,
191-203.
|
 |
|
|
|
|
 |
W.Grabarse,
M.Vaupel,
J.A.Vorholt,
S.Shima,
R.K.Thauer,
A.Wittershagen,
G.Bourenkov,
H.D.Bartunik,
and
U.Ermler
(1999).
The crystal structure of methenyltetrahydromethanopterin cyclohydrolase from the hyperthermophilic archaeon Methanopyrus kandleri.
|
| |
Structure,
7,
1257-1268.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Peisach,
D.M.Chipman,
P.W.Van Ophem,
J.M.Manning,
and
D.Ringe
(1998).
Crystallographic study of steps along the reaction pathway of D-amino acid aminotransferase.
|
| |
Biochemistry,
37,
4958-4967.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.T.Mollova,
D.E.Metzler,
A.Kintanar,
H.Kagamiyama,
H.Hayashi,
K.Hirotsu,
and
I.Miyahara
(1997).
Use of 1H-15N heteronuclear multiple-quantum coherence NMR spectroscopy to study the active site of aspartate aminotransferase.
|
| |
Biochemistry,
36,
615-625.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

