 |
PDBsum entry 1agn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1agn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
primary alcohol
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
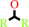 secondary alcohol
secondary alcohol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
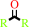 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.105
- all-trans-retinol dehydrogenase (NAD(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol--[retinol-binding protein] + NAD+ = all-trans- retinal--[retinol-binding protein] + NADH + H+
|
 |
 |
 |
 |
 |
all-trans-retinol--[retinol-binding protein]
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
all-trans- retinal--[retinol-binding protein]
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.66
- omega-hydroxydecanoate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
10-hydroxydecanoate + NAD+ = 10-oxodecanoate + NADH + H+
|
 |
 |
 |
 |
 |
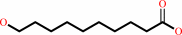 10-hydroxydecanoate
10-hydroxydecanoate
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
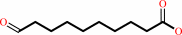 10-oxodecanoate
10-oxodecanoate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
272:18558-18563
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray structure of human class IV sigmasigma alcohol dehydrogenase. Structural basis for substrate specificity.
|
|
P.Xie,
S.H.Parsons,
D.C.Speckhard,
W.F.Bosron,
T.D.Hurley.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structural determinants of substrate recognition in the human class IV, or
sigmasigma, alcohol dehydrogenase (ADH) isoenzyme were examined through x-ray
crystallography and site-directed mutagenesis. The crystal structure of
sigmasigma ADH complexed with NAD+ and acetate was solved to 3-A resolution. The
human beta1beta1 and sigmasigma ADH isoenzymes share 69% sequence identity and
exhibit dramatically different kinetic properties. Differences in the amino
acids at positions 57, 116, 141, 309, and 317 create a different topology within
the sigmasigma substrate-binding pocket, relative to the beta1beta1 isoenzyme.
The nicotinamide ring of the NAD(H) molecule, in the sigmasigma structure,
appears to be twisted relative to its position in the beta1beta1 isoenzyme. In
conjunction with movements of Thr-48 and Phe-93, this twist widens the substrate
pocket in the vicinity of the catalytic zinc and may contribute to this
isoenzyme's high Km for small substrates. The presence of Met-57, Met-141, and
Phe-309 narrow the middle region of the sigmasigma substrate pocket and may
explain the substantially decreased Km values with increased chain length of
substrates in sigmasigma ADH. The kinetic properties of a mutant sigmasigma
enzyme (sigma309L317A) suggest that widening the middle region of the substrate
pocket increases Km by weakening the interactions between the enzyme and smaller
substrates while not affecting the binding of longer alcohols, such as hexanol
and retinol.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Comparison of   , ,   , and , and  [1] [1]  [1] dimers
by alignment of their C [1] dimers
by alignment of their C  atoms. All
C atoms. All
C  atoms were
used in the alignment. The^ result of the alignment using the CD
dimer of atoms were
used in the alignment. The^ result of the alignment using the CD
dimer of   ADH is
shown. The other dimer of ADH is
shown. The other dimer of   ADH in the
asymmetric unit gives similar results. ADH in the
asymmetric unit gives similar results.
|
 |
Figure 2.
Fig. 2. The influence of the substitution of Ala-317  -Cys in -Cys in   on the
conformation of the nicotinamide ring of NAD^+. The residues in on the
conformation of the nicotinamide ring of NAD^+. The residues in
  ADH are
shown using the thicker line. The electron density map is from
the ADH are
shown using the thicker line. The electron density map is from
the  structure
and contoured^ at 1.0 S.D. of the map. structure
and contoured^ at 1.0 S.D. of the map.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1997,
272,
18558-18563)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Ohno,
H.Hiroi,
M.Momoeda,
Y.Hosokawa,
R.Tsutsumi,
M.Koizumi,
F.Nakazawa,
T.Yano,
O.Tsutsumi,
and
Y.Taketani
(2008).
Evidence for the expression of alcohol dehydrogenase class I gene in rat uterus and its up-regulation by progesterone.
|
| |
Endocr J,
55,
83-90.
|
 |
|
|
|
|
 |
X.Parés,
J.Farrés,
N.Kedishvili,
and
G.Duester
(2008).
Medium- and short-chain dehydrogenase/reductase gene and protein families : Medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism.
|
| |
Cell Mol Life Sci,
65,
3936-3949.
|
 |
|
|
|
|
 |
C.A.Bottoms,
T.A.White,
and
J.J.Tanner
(2006).
Exploring structurally conserved solvent sites in protein families.
|
| |
Proteins,
64,
404-421.
|
 |
|
|
|
|
 |
I.Levin,
G.Meiri,
M.Peretz,
Y.Burstein,
and
F.Frolow
(2004).
The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol.
|
| |
Protein Sci,
13,
1547-1556.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Martras,
R.Alvarez,
S.E.Martínez,
D.Torres,
O.Gallego,
G.Duester,
J.Farrés,
A.R.de Lera,
and
X.Parés
(2004).
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in retina.
|
| |
Eur J Biochem,
271,
1660-1670.
|
 |
|
|
|
|
 |
E.Valencia,
A.Rosell,
C.Larroy,
J.Farrés,
J.A.Biosca,
I.Fita,
X.Parés,
and
W.F.Ochoa
(2003).
Crystallization and preliminary X-ray analysis of NADP(H)-dependent alcohol dehydrogenases from Saccharomyces cerevisiae and Rana perezi.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
334-337.
|
 |
|
|
|
|
 |
S.W.Kruse,
R.Zhao,
D.P.Smith,
and
D.N.Jones
(2003).
Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster.
|
| |
Nat Struct Biol,
10,
694-700.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Allali-Hassani,
B.Crosas,
X.Parés,
and
J.Farrés
(2001).
Kinetic effects of a single-amino acid mutation in a highly variable loop (residues 114-120) of class IV ADH.
|
| |
Chem Biol Interact,
130,
435-444.
|
 |
|
|
|
|
 |
B.V.Plapp,
J.L.Mitchell,
and
K.B.Berst
(2001).
Mouse alcohol dehydrogenase 4: kinetic mechanism, substrate specificity and simulation of effects of ethanol on retinoid metabolism.
|
| |
Chem Biol Interact,
130,
445-456.
|
 |
|
|
|
|
 |
G.Duester
(2001).
Genetic dissection of retinoid dehydrogenases.
|
| |
Chem Biol Interact,
130,
469-480.
|
 |
|
|
|
|
 |
M.S.Niederhut,
B.J.Gibbons,
S.Perez-Miller,
and
T.D.Hurley
(2001).
Three-dimensional structures of the three human class I alcohol dehydrogenases.
|
| |
Protein Sci,
10,
697-706.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Darmanin,
and
O.El-Kabbani
(2000).
Modelling studies on the binding of substrate and inhibitor to the active site of human sorbitol dehydrogenase.
|
| |
Bioorg Med Chem Lett,
10,
1101-1104.
|
 |
|
|
|
|
 |
G.Duester
(2000).
Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid.
|
| |
Eur J Biochem,
267,
4315-4324.
|
 |
|
|
|
|
 |
H.Erlandsen,
E.E.Abola,
and
R.C.Stevens
(2000).
Combining structural genomics and enzymology: completing the picture in metabolic pathways and enzyme active sites.
|
| |
Curr Opin Struct Biol,
10,
719-730.
|
 |
|
|
|
|
 |
C.Li,
J.Heatwole,
S.Soelaiman,
and
M.Shoham
(1999).
Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specificity and thermostability.
|
| |
Proteins,
37,
619-627.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Deltour,
M.H.Foglio,
and
G.Duester
(1999).
Impaired retinol utilization in Adh4 alcohol dehydrogenase mutant mice.
|
| |
Dev Genet,
25,
1.
|
 |
|
|
|
|
 |
P.T.Xie,
and
T.D.Hurley
(1999).
Methionine-141 directly influences the binding of 4-methylpyrazole in human sigma sigma alcohol dehydrogenase.
|
| |
Protein Sci,
8,
2639-2644.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Cho,
and
B.V.Plapp
(1998).
Specificity of alcohol dehydrogenases for sulfoxides.
|
| |
Biochemistry,
37,
4482-4489.
|
 |
|
|
|
|
 |
I.Hoffmann,
H.L.Ang,
and
G.Duester
(1998).
Alcohol dehydrogenases in Xenopus development: conserved expression of ADH1 and ADH4 in epithelial retinoid target tissues.
|
| |
Dev Dyn,
213,
261-270.
|
 |
|
|
|
|
 |
R.J.Haselbeck,
and
G.Duester
(1998).
ADH1 and ADH4 alcohol/retinol dehydrogenases in the developing adrenal blastema provide evidence for embryonic retinoid endocrine function.
|
| |
Dev Dyn,
213,
114-120.
|
 |
|
|
|
|
 |
S.Kotagiri,
and
H.J.Edenberg
(1998).
Regulation of human alcohol dehydrogenase gene ADH7: importance of an AP-1 site.
|
| |
DNA Cell Biol,
17,
583-590.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

