 |
PDBsum entry 1adb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase (NAD(a)-choh(d))
|
PDB id
|
|

|

|
1adb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
primary alcohol
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 66.04% similarity
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
secondary alcohol
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 66.04% similarity
|
=
|
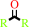 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:11734-11744
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
|
|
H.Li,
W.H.Hallows,
J.S.Punzi,
K.W.Pankiewicz,
K.A.Watanabe,
B.M.Goldstein.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CNAD (5-beta-D-ribofuranosylnicotinamide adenine dinucleotide) is an isosteric
C-glycosidic analogue of NAD(H) containing a neutral pyridine ring. CPAD
(5-beta-D-ribofuranosylpicolinamide adenine dinucleotide) is a closely related
pyridine-containing analogue with the pyridine nitrogen on the opposite side of
the ring. CNAD is a potent and specific inhibitor of horse liver alcohol
dehydrogenase (LADH), binding with a dissociation constant in the nanomolar
range. CPAD binds LADH with an affinity comparable to that of NAD. Crystal
structures of CNAD and CPAD bound to LADH are presented at 2.4 and 2.7 A,
respectively. The two complexes are isomorphous, crystallizing in the triclinic
system with cell dimensions different from those seen in previous ternary LADH
complexes. Structures were solved using the molecular replacement method and
refined to crystallographic R values of 18% (CNAD) and 17% (CPAD). Both
inhibitors bind to the "closed" form of LADH in the normal cofactor-binding
cleft. The conformation of LADH-bound CPAD closely mimics that of LADH-bound
NAD(H). The data suggest that alcohol substrate binds directly to the catalytic
zinc atom. In the CNAD complex, the pyridine nitrogen replaces alcohol as the
fourth coordination ligand to the active site zinc atom, while all other polar
interactions remain the same as those of bound NAD(H). The zinc-nitrogen ligand
explains the high affinity of CNAD for LADH.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.V.Plapp
(2010).
Conformational changes and catalysis by alcohol dehydrogenase.
|
| |
Arch Biochem Biophys,
493,
3.
|
 |
|
|
|
|
 |
D.Coquière,
S.Le Gac,
U.Darbost,
O.Sénèque,
I.Jabin,
and
O.Reinaud
(2009).
Biomimetic and self-assembled calix[6]arene-based receptors for neutral molecules.
|
| |
Org Biomol Chem,
7,
2485-2500.
|
 |
|
|
|
|
 |
C.C.Milburn,
H.J.Lamble,
A.Theodossis,
S.D.Bull,
D.W.Hough,
M.J.Danson,
and
G.L.Taylor
(2006).
The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
|
| |
J Biol Chem,
281,
14796-14804.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Hayward,
and
A.Kitao
(2006).
Molecular dynamics simulations of NAD+-induced domain closure in horse liver alcohol dehydrogenase.
|
| |
Biophys J,
91,
1823-1831.
|
 |
|
|
|
|
 |
A.Tousignant,
and
J.N.Pelletier
(2004).
Protein motions promote catalysis.
|
| |
Chem Biol,
11,
1037-1042.
|
 |
|
|
|
|
 |
L.Esposito,
I.Bruno,
F.Sica,
C.A.Raia,
A.Giordano,
M.Rossi,
L.Mazzarella,
and
A.Zagari
(2003).
Crystal structure of a ternary complex of the alcohol dehydrogenase from Sulfolobus solfataricus.
|
| |
Biochemistry,
42,
14397-14407.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.M.Goldstein,
and
T.D.Colby
(2000).
Conformational constraints in NAD analogs: implications for dehydrogenase binding and specificity.
|
| |
Adv Enzyme Regul,
40,
405-426.
|
 |
|
|
|
|
 |
H.W.Adolph,
P.Zwart,
R.Meijers,
I.Hubatsch,
M.Kiefer,
V.Lamzin,
and
E.Cedergren-Zeppezauer
(2000).
Structural basis for substrate specificity differences of horse liver alcohol dehydrogenase isozymes.
|
| |
Biochemistry,
39,
12885-12897.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.J.Ansell,
D.A.Small,
and
C.R.Lowe
(1999).
Synthesis and properties of new coenzyme mimics based on the artificial coenzyme CL4.
|
| |
J Mol Recognit,
12,
45-56.
|
 |
|
|
|
|
 |
T.D.Colby,
B.J.Bahnson,
J.K.Chin,
J.P.Klinman,
and
B.M.Goldstein
(1998).
Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes.
|
| |
Biochemistry,
37,
9295-9304.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.W.Adolph,
M.Kiefer,
and
E.Cedergren-Zeppezauer
(1997).
Electrostatic effects in the kinetics of coenzyme binding to isozymes of alcohol dehydrogenase from horse liver.
|
| |
Biochemistry,
36,
8743-8754.
|
 |
|
|
|
|
 |
K.W.Pankiewicz
(1997).
Novel nicotinamide adenine dinucleotide analogues as potential anticancer agents: quest for specific inhibition of inosine monophosphate dehydrogenase.
|
| |
Pharmacol Ther,
76,
89.
|
 |
|
|
|
|
 |
L.Hemmingsen,
R.Bauer,
M.J.Bjerrum,
H.W.Adolph,
M.Zeppezauer,
and
E.Cedergren-Zeppezauer
(1996).
The protein conformation of Cd-substituted horse liver alcohol dehydrogenase and its metal-site coordination geometry in binary and ternary inhibitor complexes.
|
| |
Eur J Biochem,
241,
546-551.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

