- Course overview
- Search within this course
- What is UniProt?
- Why do we need UniProt?
- When to use UniProt
- Quiz: Check your learning I
- How to access and navigate UniProt
- How to search UniProt
- Annotation score
- Quiz: Check your learning II
- Exploring a UniProtKB entry
- How to use UniProt tools
- How to get data from UniProt
- How to submit data to UniProt
- When to use UniProt: guided examples
- Exercise: finding entries with 3D structures
- Exercise: mapping other database identifiers to UniProt
- Summary
- Get help and support on UniProt
- References
- Next steps
- Your feedback
Sequence data
UniProtKB sequences
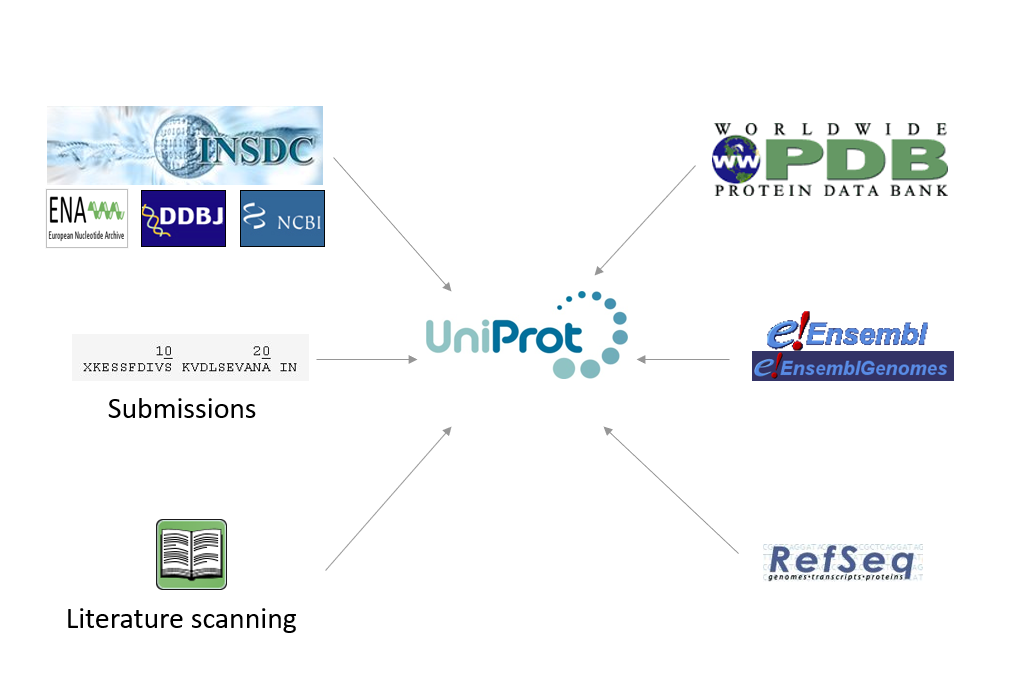
Most of the protein sequences provided by UniProtKB come from the translations of coding sequences (CDS) submitted to the ENA/GenBank/DDBJ nucleotide sequence resources of the International Nucleotide Sequence Database Collaboration (INSDC). These CDS are either generated by gene prediction programs or are experimentally proven. The translated CDS sequences are automatically transferred to the TrEMBL section of UniProtKB. The UniProtKB/TrEMBL records may eventually be selected for manual annotation and then integrated into the UniProtKB/Swiss-Prot section.
In addition to translated CDS, UniProtKB protein sequences may come from:
- The PDB database of protein structures
- Sequences experimentally obtained by direct protein sequencing and submitted to UniProt
- Sequences scanned from the literature
- Sequences derived from gene prediction but which have not been submitted to ENA/GenBank/DDBJ. These are imported from resources such as Ensembl and RefSeq
Importing and combining sequences from a range of sources means that UniProt provides a complete collection of protein sequences and contributes to consistency of protein sets across various sequence resources (Figure 3).